Abstract
Microsatellite instability (MSI) testing has been advocated for all newly diagnosed colorectal cancer patients. One of the most common tests is composed by a pentaplex panel of mononucleotides markers (NR-27, NR-21, NR-24, BAT-25, and BAT-26), which allows the analysis of MSI in tumors without the need of reference DNA. For that, it is fundamental to establish a quasi-monomorphic variation range (QMVR) for each marker. Herein, we aimed to establish the QMVR in a Brazilian healthy population, to evaluate the feasibility of MSI determination of tumors, without the matching normal DNA. Furthermore, we intend to assess their ancestry using specific ancestry-informative markers (AIMs) and correlate with QMVR. The QMVR was assessed in 214 individuals, through a pentaplex PCR followed by fragment analysis. The ancestry analysis was done by 46 AIMs in a single multiplex PCR followed by capillary electrophoresis. Following QMVR establishment, we observed 23 individuals with alleles outside the QMVR. Importantly, none of them exhibited more than one marker outside the range. Therefore, individuals with instability at ≥2 markers would be accurately classified as MSI. The European ancestry proportion was the most frequent (67.5%), followed by the African (19.6%). The comparison of the individuals with alleles within (n=191) and outside (n=23) the QMVR showed statistical difference in the proportions of European and African alleles, confirming the higher polymorphic nature of African ancestry. In conclusion, the present study reports an accurate methodology to assess MSI status without matched-normal DNA and independently of the ethnicity, even in the highly admixed population of Brazil.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Karran P : Microsatellite instability and DNA mismatch repair in human cancer. Semin Cancer Biol 1996; 7: 15–24.
Vilar E, Gruber SB : Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010; 7: 153–162.
Boland CR, Thibodeau SN, Hamilton SR et al: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998; 58: 5248–5257.
Viana-Pereira M, Lee A, Popov S et al: Microsatellite instability in pediatric high grade glioma is associated with genomic profile and differential target gene inactivation. PLoS One 2011; 6: e20588.
Boland CR, Goel A : Microsatellite instability in colorectal cancer. Gastroenterology 2010; 138: 2073–2087, e2073.
Thibodeau SN, Bren G, Schaid D : Microsatellite instability in cancer of the proximal colon. Science 1993; 260: 816–819.
Vilkki S, Launonen V, Karhu A, Sistonen P, Vastrik I, Aaltonen LA : Screening for microsatellite instability target genes in colorectal cancers. J Med Genet 2002; 39: 785–789.
Suraweera N, Duval A, Reperant M et al: Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 2002; 123: 1804–1811.
Fearon ER : Molecular genetics of colorectal cancer. Annu Rev Pathol 2011; 6: 479–507.
Calin GA, Gafa R, Tibiletti MG et al: Genetic progression in microsatellite instability high (MSI-H) colon cancers correlates with clinico-pathological parameters: a study of the TGRbetaRII, BAX, hMSH3, hMSH6, IGFIIR and BLM genes. Int J Cancer 2000; 89: 230–235.
Jass JR, Do KA, Simms LA et al: Morphology of sporadic colorectal cancer with DNA replication errors. Gut 1998; 42: 673–679.
Ribic CM, Sargent DJ, Moore MJ et al: Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003; 349: 247–257.
Sinicrope FA, Sargent DJ : Molecular pathways: microsatellite instability in colorectal cancer: prognostic, predictive, and therapeutic implications. Clin Cancer Res 2012; 18: 1506–1512.
Vilar E, Tabernero J : Molecular dissection of microsatellite instable colorectal cancer. Cancer Discov 2013; 3: 502–511.
Dorard C, de Thonel A, Collura A et al: Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med 2011; 17: 1283–1289.
Alhopuro P, Sammalkorpi H, Niittymaki I et al: Candidate driver genes in microsatellite-unstable colorectal cancer. Int J Cancer 2012; 130: 1558–1566.
Mori Y, Yin J, Rashid A et al: Instabilotyping: comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res 2001; 61: 6046–6049.
Oliveira C, Pinto M, Duval A et al: BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene 2003; 22: 9192–9196.
Parsons DW, Wang TL, Samuels Y et al: Colorectal cancer: mutations in a signalling pathway. Nature 2005; 436: 792.
Goel A, Arnold CN, Niedzwiecki D et al: Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res 2004; 64: 3014–3021.
Koopman M, Kortman GA, Mekenkamp L et al: Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 2009; 100: 266–273.
Roth AD, Tejpar S, Delorenzi M et al: Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010; 28: 466–474.
Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B : Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet 2000; 355: 1745–1750.
Sargent DJ, Marsoni S, Monges G et al: Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 2010; 28: 3219–3226.
Sinicrope FA, Foster NR, Thibodeau SN et al: DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst 2011; 103: 863–875.
Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B : Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer 2009; 45: 1890–1896.
Umar A, Boland CR, Terdiman JP et al: Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004; 96: 261–268.
Buhard O, Cattaneo F, Wong YF et al: Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol 2006; 24: 241–251.
Goel A, Nagasaka T, Hamelin R, Boland CR : An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS ONE 2010; 5: e9393.
Patil DT, Bronner MP, Portier BP, Fraser CR, Plesec TP, Liu X : A five-marker panel in a multiplex PCR accurately detects microsatellite instability-high colorectal tumors without control DNA. Diagn Mol Pathol 2012; 21: 127–133.
Viana-Pereira M, Almeida I, Sousa S et al: Analysis of microsatellite instability in medulloblastoma. Neuro Oncol 2009; 11: 458–467.
Xicola RM, Llor X, Pons E et al: Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst 2007; 99: 244–252.
Pereira R, Phillips C, Pinto N et al: Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS One 2012; 7: e29684.
Parra EJ, Marcini A, Akey J et al: Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 1998; 63: 1839–1851.
Kosoy R, Nassir R, Tian C et al: Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 2009; 30: 69–78.
Tian C, Gregersen PK, Seldin MF : Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet 2008; 17: R143–R150.
Manta FS, Pereira R, Caiafa A, Silva DA, Gusmão L, Carvalho EF : Analysis of genetic ancestry in the admixed Brazilian population from Rio de Janeiro using 46 autosomal ancestry-informative indel markers. Ann Hum Biol 2013; 40: 94–98.
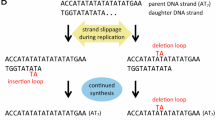
Buhard O, Suraweera N, Lectard A, Duval A, Hamelin R : Quasimonomorphic mononucleotide repeats for high-level microsatellite instability analysis. Dis Markers 2004; 20: 251–257.
Pritchard JK, Stephens M, Donnelly P : Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–959.
Falush D, Stephens M, Pritchard JK : Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003; 164: 1567–1587.
Berg AO, Armstrong K, Botkin J et al: Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 2009; 11: 35–41.
Pena SD, Di Pietro G, Fuchshuber-Moraes M et al: The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One 2011; 6: e17063.
Acknowledgements
The present study was partially supported by a MCT/FINEP/CT-INFRA-PROINFRA 02/2010 grant and a CNPq Universal grant (475358/2011-2). Nathália Cristina Campanella is a recipient of an FAPESP Master Grant (2012/01732-2). IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by the Portuguese Foundation for Science and Technology (FCT). Rui Pereira is a recipient of a postdoctoral fellowship from FCT (SFRH/BPD/81986/2011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Campanella, N., Berardinelli, G., Scapulatempo-Neto, C. et al. Optimization of a pentaplex panel for MSI analysis without control DNA in a Brazilian population: correlation with ancestry markers. Eur J Hum Genet 22, 875–880 (2014). https://doi.org/10.1038/ejhg.2013.256
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.256
Keywords
This article is cited by
-
Association of microsatellite instability (MSI) status with the 5-year outcome and genetic ancestry in a large Brazilian cohort of colorectal cancer
European Journal of Human Genetics (2022)
-
Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations
Scientific Reports (2019)
-
Universal determination of microsatellite instability using BAT26 as a single marker in an Argentine colorectal cancer cohort
Familial Cancer (2018)