Abstract
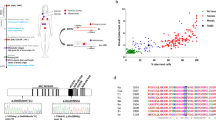
Fanconi anaemia (FA) is characterized by progressive bone marrow failure, congenital anomalies, and predisposition to malignancy. In a minority of cases, FA results from biallelic FANCD1/BRCA2 mutations that are associated with early-onset leukaemia and solid tumours. Here, we describe the clinical and molecular features of a remarkable family presenting with multiple primary colorectal cancers (CRCs) without detectable mutations in genes involved in the Mendelian predisposition to CRCs. We unexpectedly identified, despite the absence of clinical cardinal features of FA, a biallelic mutation of the FANCD1/BRCA2 corresponding to a frameshift alteration (c.1845_1846delCT, p.Asn615Lysfs*6) and a missense mutation (c.7802A>G, p.Tyr2601Cys). The diagnosis of FA was confirmed by the chromosomal analysis of lymphocytes. Reverse transcriptase (RT)-PCR analysis revealed that the c.7802A>G BRCA2 variation was in fact a splicing mutation that creates an aberrant splicing donor site and results partly into an aberrant transcript encoding a truncated protein (p.Tyr2601Trpfs*46). The atypical FA phenotype observed within this family was probably explained by the residual amount of BRCA2 with the point mutation c.7802A>G in the patients harbouring the biallelic FANCD1/BRCA2 mutations. Although this report is based in a single family, it suggests that CRCs may be part of the tumour spectrum associated with FANCD1/BRCA2 biallelic mutations and that the presence of such mutations should be considered in families with CRCs, even in the absence of cardinal features of FA.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Joenje H, Patel KJ : The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet 2001; 2: 446–457.
Kutler DI, Singh B, Satagopan J et al: A 20-year perspective on the International Fanconi Anaemia Registry (IFAR). Blood 2003; 101: 1249–1256.
Tischkowitz MD, Hodgson SV : Fanconi anaemia. J Med Genet 2003; 40: 1–10.
de Winter JP, Joenje H : The genetic and molecular basis of Fanconi anaemia. Mutat Res 2009; 668: 11–19.
Levitus M, Joenje H, de Winter JP : The Fanconi anaemia pathway of genomic maintenance. Cell Oncol 2006; 28: 3–29.
Schroeder TM, Anschutz F, Knopp A : [Spontaneous chromosome aberrations in familial panmyelopathy]. Humangenetik 1964; 1: 194–196.
Sasaki MS, Tonomura A : A high susceptibility of Fanconi's anaemia to chromosome breakage by DNA cross-linking agents. Cancer Res 1973; 33: 1829–1836.
Rosenberg PS, Greene MH, Alter BP : Cancer incidence in persons with Fanconi anaemia. Blood 2003; 101: 822–826.
Shimamura A, Alter BP : Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev 2010; 24: 101–122.
Alter BP : Cancer in Fanconi anaemia, 1927-2001. Cancer 2003; 97: 425–440.
Rosenberg PS, Huang Y, Alter BP : Individualized risks of first adverse events in patients with Fanconi anaemia. Blood 2004; 104: 350–355.
Alter BP, Rosenberg PS, Brody LC : Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet 2007; 44: 1–9.
Reese MG, Eeckman FH, Kulp D, Haussler D : Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323.
Shapiro MB, Senapathy P : RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 1987; 15: 7155–7174.
Yeo G, Burge CB : Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 2004; 11: 377–394.
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR : ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003; 31: 3568–3571.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C : Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67.
Neveling K, Endt D, Hoehn H, Schindler D : Genotype-phenotype correlations in Fanconi anaemia. Mutat Res 2009; 668: 73–91.
Faivre L, Portnoi MF, Pals G et al: Should chromosome breakage studies be performed in patients with VACTERL association? Am J Med Genet A 2005; 137: 55–58.
Hirsch B, Shimamura A, Moreau L et al: Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood 2004; 103: 2554–2559.
Howlett NG, Taniguchi T, Olson S et al: Biallelic inactivation of BRCA2 in Fanconi anaemia. Science 2002; 297: 606–609.
Wagner JE, Tolar J, Levran O et al: Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukaemia, and Fanconi anaemia. Blood 2004; 103: 3226–3229.
Dewire MD, Ellison DW, Patay Z, McKinnon PJ, Sanders RP, Gajjar A : Fanconi anaemia and biallelic BRCA2 mutation diagnosed in a young child with an embryonal CNS tumour. Pediatr Blood Cancer 2009; 53: 1140–1142.
Ikeda H, Matsushita M, Waisfisz Q et al: Genetic reversion in an acute myelogenous leukaemia cell line from a Fanconi anaemia patient with biallelic mutations in BRCA2. Cancer Res 2003; 63: 2688–2694.
Meyer S, Fergusson WD, Oostra AB et al: A cross-linker-sensitive myeloid leukaemia cell line from a 2-year-old boy with severe Fanconi anaemia and biallelic FANCD1/BRCA2 mutations. Genes Chromosomes Cancer 2005; 42: 404–415.
Myers K, Davies SM, Harris RE et al: The clinical phenotype of children with Fanconi anaemia caused by biallelic FANCD1/BRCA2 mutations. Pediatr Blood Cancer 2012; 58: 462–465.
Offit K, Levran O, Mullaney B et al: Shared genetic susceptibility to breast cancer, brain tumors, and Fanconi anaemia. J Natl Cancer Inst 2003; 95: 1548–1551.
Reid S, Renwick A, Seal S et al: Biallelic BRCA2 mutations are associated with multiple malignancies in childhood including familial Wilms tumour. J Med Genet 2005; 42: 147–151.
Rahman N, Scott RH : Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum Mol Genet 2007; 16 Spec No 1: R60–R66.
Cartegni L, Chew SL, Krainer AR : Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 2002; 3: 285–298.
Matsuda D, Sato H, Maquat LE : Chapter 9. Studying nonsense-mediated mRNA decay in mammalian cells. Methods Enzymol 2008; 449: 177–201.
Grady WM, Markowitz SD : Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet 2002; 3: 101–128.
Muller A, Fishel R : Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC). Cancer Invest 2002; 20: 102–109.
Peng M, Litman R, Xie J, Sharma S, Brosh RM Jr., Cantor SB : The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J 2007; 26: 3238–3249.
Williams SA, Wilson JB, Clark AP et al: Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet 2011; 20: 4395–4410.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author contributions
M-MP, SG, FC, CL, CH, EF, AD, GC, SEC, PF, LF, AB, CL, J-LJ provided clinical data. ED-C, JS, MB, CC, CC, LM, SL, LA, PC, FM, TF, PJ and LF contributed to conception and design, acquisition of data, and analysis and interpretation of data. ED-C, JS, TF, PJ, LF contributed to writing the manuscript.
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Degrolard-Courcet, E., Sokolowska, J., Padeano, MM. et al. Development of primary early-onset colorectal cancers due to biallelic mutations of the FANCD1/BRCA2 gene. Eur J Hum Genet 22, 979–987 (2014). https://doi.org/10.1038/ejhg.2013.278
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.278
Keywords
This article is cited by
-
Clinical consequences of BRCA2 hypomorphism
npj Breast Cancer (2021)