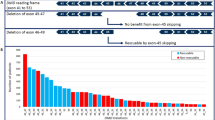
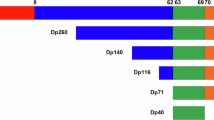
Abstract
Duchenne and Becker muscular dystrophies (DMD/BMD) are the most commonly inherited neuromuscular disease. However, accurate and convenient molecular diagnosis cannot be achieved easily because of the enormous size of the dystrophin gene and complex causative mutation spectrum. Such traditional methods as multiplex ligation-dependent probe amplification plus Sanger sequencing require multiple steps to fulfill the diagnosis of DMD/BMD. Here, we introduce a new single-step method for the genetic analysis of DMD patients and female carriers in real clinical settings and demonstrate the validation of its accuracy. A total of 89 patients, 18 female carriers and 245 non-DMD patients were evaluated using our targeted NGS approaches. Compared with traditional methods, our new method yielded 99.99% specificity and 98.96% sensitivity for copy number variations detection and 100% accuracy for the identification of single-nucleotide variation mutations. Additionally, this method is able to detect partial deletions/duplications, thus offering precise personal DMD gene information for gene therapy. We detected novel partial deletions of exons in nine samples for which the breakpoints were located within exonic regions. The results proved that our new method is suitable for routine clinical practice, with shorter turnaround time, higher accuracy, and better insight into comprehensive genetic information (detailed breakpoints) for ensuing gene therapy.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bushby K, Finkel R, Birnkrant DJ et al: Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol 2010; 9: 77–93.
Hoffman EP, Brown RH, Kunkel LM : Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987; 51: 919–928.
Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT : Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 2006; 34: 135–144.
Lalic T, Vossen RH, Coffa J et al: Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet 2005; 13: 1231–1234.
Hegde MR, Chin EL, Mulle JG, Okou DT, Warren ST, Zwick ME : Microarray-based mutation detection in the dystrophin gene. Hum Mutat 2008; 29: 1091–1099.
Bennett RR, den Dunnen J, O'Brien KF, Darras BT, Kunkel LM : Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genet 2001; 2: 17.
Flanigan KM, Dunn DM, von Niederhausern A et al: Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 2009; 30: 1657–1666.
Pichavant C, Aartsma-Rus A, Clemens PR et al: Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol Ther 2011; 19: 830–840.
Malik V, Rodino-Klapac LR, Viollet L et al: Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol 2010; 67: 771–780.
Welch EM, Barton ER, Zhuo J et al: PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007; 447: 87–91.
Cirak S, Arechavala-Gomeza V, Guglieri M et al: Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet 2011; 378: 595–605.
Goemans NM, Tulinius M, van den Akker JT et al: Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med 2011; 364: 1513–1522.
Ng SB, Buckingham KJ, Lee C et al: Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010; 42: 30–35.
Calvo SE, Compton AG, Hershman SG et al: Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med 2012; 4: 118ra110.
Lim BC, Lee S, Shin JY et al: Genetic diagnosis of Duchenne and Becker muscular dystrophy using next-generation sequencing technology: comprehensive mutational search in a single platform. J Med Genet 2011; 48: 731–736.
Kunkel LM, Hejtmancik JF, Caskey CT et al: Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature 1986; 322: 73–77.
Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT : Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 1988; 16: 11141–11156.
del Gaudio D, Yang Y, Boggs BA et al: Molecular diagnosis of Duchenne/Becker muscular dystrophy: enhanced detection of dystrophin gene rearrangements by oligonucleotide array-comparative genomic hybridization. Hum Mutat 2008; 29: 1100–1107.
Hall N : Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol 2007; 210: 1518–1525.
Xie S, Lan Z, Qu N et al: Detection of truncated dystrophin lacking the C-terminal domain in a Chinese pedigree by next-generation sequencing. Gene 2012; 499: 139–142.
Chiu RW, Akolekar R, Zheng YW et al: Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 2011; 342: c7401.
Bell CJ, Dinwiddie DL, Miller NA et al: Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 2011; 3: 65ra64.
Politano L, Nigro V, Nigro G et al: Development of cardiomyopathy in female carriers of Duchenne and Becker muscular dystrophies. JAMA 1996; 275: 1335–1338.
Mendell JR, Shilling C, Leslie ND et al: Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol 2012; 71: 304–313.
Loman NJ, Misra RV, Dallman TJ et al: Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012; 30: 434–439.
Bovolenta M, Scotton C, Falzarano MS, Gualandi F, Ferlini A : Rapid, comprehensive analysis of the dystrophin transcript by a custom micro-fluidic exome array. Hum Mutat 2012; 33: 572–581.
Acknowledgements
We thank all the blood donors for their invaluable contribution to this study. The study received financial support from Shenzhen Development and Reform Commission project, Guangdong Provincial Science and Technology Department Planning Project, Shenzhen Birth Defect Screening Project Lab (JZF No. (2011) 861) and Peking Union Medical College Hospital Young Scholars Project. Both of the projects are non-profit research projects by government and institutions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Wei, X., Dai, Y., Yu, P. et al. Targeted next-generation sequencing as a comprehensive test for patients with and female carriers of DMD/BMD: a multi-population diagnostic study. Eur J Hum Genet 22, 110–118 (2014). https://doi.org/10.1038/ejhg.2013.82
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.82
Keywords
This article is cited by
-
Identification of two novel large deletions in FBN1 gene by next-generation sequencing and multiplex ligation-dependent probe amplification
BMC Medical Genomics (2024)
-
A female patient carrying a novel DMD mutation with non-random X-chromosome inactivation from a DMD family
BMC Medical Genomics (2024)
-
DMD/BMD prenatal diagnosis and treatment expectation in a single centre in China for 15 years
BMC Medical Genomics (2021)
-
Mutation Spectrum of Dystrophinopathies in India: Implications for Therapy
The Indian Journal of Pediatrics (2020)
-
Genetic analysis of 62 Chinese families with Duchenne muscular dystrophy and strategies of prenatal diagnosis in a single center
BMC Medical Genetics (2019)