Abstract
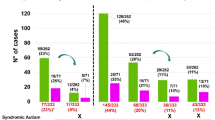
Copy number variants (CNVs) have repeatedly been found to cause or predispose to autism spectrum disorders (ASDs). For diagnostic purposes, we screened 194 individuals with ASDs for CNVs using Illumina SNP arrays. In several probands, we also analyzed candidate genes located in inherited deletions to unmask autosomal recessive variants. Three CNVs, a de novo triplication of chromosome 15q11–q12 of paternal origin, a deletion on chromosome 9p24 and a de novo 3q29 deletion, were identified as the cause of the disorder in one individual each. An autosomal recessive cause was considered possible in two patients: a homozygous 1p31.1 deletion encompassing PTGER3 and a deletion of the entire DOCK10 gene associated with a rare hemizygous missense variant. We also identified multiple private or recurrent CNVs, the majority of which were inherited from asymptomatic parents. Although highly penetrant CNVs or variants inherited in an autosomal recessive manner were detected in rare cases, our results mainly support the hypothesis that most CNVs contribute to ASDs in association with other CNVs or point variants located elsewhere in the genome. Identification of these genetic interactions in individuals with ASDs constitutes a formidable challenge.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Miles JH : Autism spectrum disorders – a genetics review. Genet Med 2011; 13: 278–294.
Ozonoff S, Young GS, Carter A et al: Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics 2011; 128: e488–e495.
Glessner JT, Wang K, Cai G et al: Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009; 459: 569–573.
Bucan M, Abrahams BS, Wang K et al: Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet 2009; 5: e1000536.
Sanders SJ, Ercan-Sencicek AG, Hus V et al: Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 2011; 70: 863–885.
O’Roak BJ, Vives L, Girirajan S et al: Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 246–250.
Sanders SJ, Murtha MT, Gupta AR et al: De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012; 485: 237–241.
Neale BM, Kou Y, Liu L et al: Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 242–245.
Szatmari P, Paterson AD, Zwaigenbaum L et al: Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 2007; 39: 319–328.
Schaaf CP, Zoghbi HY : Solving the autism puzzle a few pieces at a time. Neuron 2011; 70: 806–808.
Levy D, Ronemus M, Yamrom B et al: Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 2011; 70: 886–897.
Guilmatre A, Dubourg C, Mosca AL et al: Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry 2009; 66: 947–956.
Depienne C, Moreno-De-Luca D, Heron D et al: Screening for genomic rearrangements and methylation abnormalities of the 15q11–q13 region in autism spectrum disorders. Biol Psychiatry 2009; 66: 349–359.
Gai X, Xie HM, Perin JC et al: Rare structural variation of synapse and neurotransmission genes in autism. Mol Psychiatry 2012; 17: 402–411.
Jamain S, Quach H, Betancur C et al: Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 2003; 34: 27–29.
Durand CM, Betancur C, Boeckers TM et al: Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 2007; 39: 25–27.
Leblond CS, Heinrich J, Delorme R et al: Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet 2012; 8: e1002521.
Betancur C : Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res 2011; 1380: 42–77.
Schaaf CP, Sabo A, Sakai Y et al: Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet 2011; 20: 3366–3375.
Basu SN, Kollu R, Banerjee-Basu S : AutDB: a gene reference resource for autism research. Nucleic Acids Res 2009; 37: D832–D836.
Hochstenbach R, Poot M, Nijman IJ et al: Discovery of variants unmasked by hemizygous deletions. Eur J Hum Genet 2012; 20: 748–753.
Newbury DF, Winchester L, Addis L et al: CMIP and ATP2C2 modulate phonological short-term memory in language impairment. Am J Hum Genet 2009; 85: 264–272.
Salyakina D, Cukier HN, Lee JM et al: Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PLoS One 2011; 6: e26049.
van Daalen E, Kemner C, Verbeek NE et al: Social Responsiveness Scale-aided analysis of the clinical impact of copy number variations in autism. Neurogenetics 2011; 12: 315–323.
Veltman MW, Thompson RJ, Craig EE et al: A paternally inherited duplication in the Prader–Willi/Angelman syndrome critical region: a case and family study. J Autism Dev Disord 2005; 35: 117–127.
Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM : 15q11–13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet 2007; 16: 691–703.
Vinci G, Chantot-Bastaraud S, El Houate B, Lortat-Jacob S, Brauner R, McElreavey K : Association of deletion 9p, 46,XY gonadal dysgenesis and autistic spectrum disorder. Mol Hum Reprod 2007; 13: 685–689.
Sakai Y, Shaw CA, Dawson BC et al: Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med 2011; 3: 86ra49.
Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D : Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011; 70: 898–907.
Van Houdt JK, Nowakowska BA, Sousa SB et al: Heterozygous missense mutations in SMARCA2 cause Nicolaides–Baraitser syndrome. Nat Genet 2012; 44: 445–449, S441.
Willatt L, Cox J, Barber J et al: 3q29 Microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet 2005; 77: 154–160.
Quintero-Rivera F, Sharifi-Hannauer P, Martinez-Agosto JA : Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: case report and review. Am J Med Genet A 2010; 152A: 2459–2467.
Mulle JG, Dodd AF, McGrath JA et al: Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet 2010; 87: 229–236.
Cobb W, Anderson A, Turner C, Hoffman RD, Schonberg S, Levin SW : 1.3 Mb de novo deletion in chromosome band 3q29 associated with normal intelligence in a child. Eur J Med Genet 2010; 53: 415–418.
Li F, Lisi EC, Wohler ES, Hamosh A, Batista DA : 3q29 interstitial microdeletion syndrome: an inherited case associated with cardiac defect and normal cognition. Eur J Med Genet 2009; 52: 349–352.
Weiss LA, Shen Y, Korn JM et al: Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 2008; 358: 667–675.
Sahoo T, Theisen A, Rosenfeld JA et al: Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med 2011; 13: 868–880.
Cooper GM, Coe BP, Girirajan S et al: A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–846.
Maestrini E, Pagnamenta AT, Lamb JA et al: High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry 2010; 15: 954–968.
Momiyama T, Todo N, Sugimoto Y, Ichikawa A, Narumiya S : Membrane depolarization by activation of prostaglandin E receptor EP3 subtype of putative serotonergic neurons in the dorsal raphe nucleus of the rat. Naunyn Schmiedebergs Arch Pharmacol 1996; 353: 377–381.
Ushikubi F, Segi E, Sugimoto Y et al: Impaired febrile response in mice lacking the prostaglandin E receptor subtype EP3. Nature 1998; 395: 281–284.
Matsuoka Y, Furuyashiki T, Bito H et al: Impaired adrenocorticotropic hormone response to bacterial endotoxin in mice deficient in prostaglandin E receptor EP1 and EP3 subtypes. Proc Natl Acad Sci USA 2003; 100: 4132–4137.
Kunikata T, Yamane H, Segi E et al: Suppression of allergic inflammation by the prostaglandin E receptor subtype EP3. Nat Immunol 2005; 6: 524–531.
Sanchez-Alavez M, Klein I, Brownell SE et al: Night eating and obesity in the EP3R-deficient mouse. Proc Natl Acad Sci USA 2007; 104: 3009–3014.
Aronoff DM, Lewis C, Serezani CH et al: E-prostanoid 3 receptor deletion improves pulmonary host defense and protects mice from death in severe Streptococcus pneumoniae infection. J Immunol 2009; 183: 2642–2649.
Sugimoto Y, Narumiya S : Prostaglandin E receptors. J Biol Chem 2007; 282: 11613–11617.
Gau SS, Liao HM, Hong CC, Chien WH, Chen CH : Identification of two inherited copy number variants in a male with autism supports two-hit and compound heterozygosity models of autism. Am J Med Genet B 2012; 159B: 710–717.
Guinchat V, Thorsen P, Laurent C, Cans C, Bodeau N, Cohen D : Pre-, peri- and neonatal risk factors for autism. Acta Obstet Gynecol Scand 2012; 91: 287–300.
Acknowledgements
We thank the P3S platform, the ‘Plateau technique Mutualisé du GHU Est’, and the genotyping and sequencing platform of the ICM for technical assistance and the DNA and cell bank of CRICM for DNA extraction and cell culture. We also thank Dr Merle Ruberg for critical reading of the manuscript and Sophie Rivaud-Pechoux for her help with the statistical analysis. This study was financially supported by AP-HP (DHOS), Fondation de France, ERA-NET NEURON EUHFAUTISM and INSERM.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Nava, C., Keren, B., Mignot, C. et al. Prospective diagnostic analysis of copy number variants using SNP microarrays in individuals with autism spectrum disorders. Eur J Hum Genet 22, 71–78 (2014). https://doi.org/10.1038/ejhg.2013.88
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.88
Keywords
This article is cited by
-
Identification of primary copy number variations reveal enrichment of Calcium, and MAPK pathways sensitizing secondary sites for autism
Egyptian Journal of Medical Human Genetics (2020)
-
PRMT7 deficiency causes dysregulation of the HCN channels in the CA1 pyramidal cells and impairment of social behaviors
Experimental & Molecular Medicine (2020)
-
Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders
Cell Research (2018)
-
Chromosomal microarray analysis in a cohort of underrepresented population identifies SERINC2 as a novel candidate gene for autism spectrum disorder
Scientific Reports (2017)
-
Overexpression of Homer1a in the basal and lateral amygdala impairs fear conditioning and induces an autism-like social impairment
Molecular Autism (2016)