Abstract
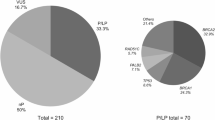
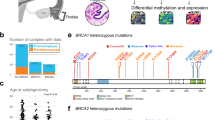
To optimize the molecular diagnosis of hereditary breast and ovarian cancer (HBOC), we developed a next-generation sequencing (NGS)-based screening based on the capture of a panel of genes involved, or suspected to be involved in HBOC, on pooling of indexed DNA and on paired-end sequencing in an Illumina GAIIx platform, followed by confirmation by Sanger sequencing or MLPA/QMPSF. The bioinformatic pipeline included CASAVA, NextGENe, CNVseq and Alamut-HT. We validated this procedure by the analysis of 59 patients’ DNAs harbouring SNVs, indels or large genomic rearrangements of BRCA1 or BRCA2. We also conducted a blind study in 168 patients comparing NGS versus Sanger sequencing or MLPA analyses of BRCA1 and BRCA2. All mutations detected by conventional procedures were detected by NGS. We then screened, using three different versions of the capture set, a large series of 708 consecutive patients. We detected in these patients 69 germline deleterious alterations within BRCA1 and BRCA2, and 4 TP53 mutations in 468 patients also tested for this gene. We also found 36 variations inducing either a premature codon stop or a splicing defect among other genes: 5/708 in CHEK2, 3/708 in RAD51C, 1/708 in RAD50, 7/708 in PALB2, 3/708 in MRE11A, 5/708 in ATM, 3/708 in NBS1, 1/708 in CDH1, 3/468 in MSH2, 2/468 in PMS2, 1/708 in BARD1, 1/468 in PMS1 and 1/468 in MLH3. These results demonstrate the efficiency of NGS in performing molecular diagnosis of HBOC. Detection of mutations within other genes than BRCA1 and BRCA2 highlights the genetic heterogeneity of HBOC.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Miki Y, Swensen J, Shattuck-Eidens D et al: A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266: 66–71.
Wooster R, Bignell G, Lancaster J et al: Identification of the breast cancer susceptibility gene BRCA2. Nature 1995; 378: 789–792.
Antoniou A, Pharoah PD, Narod S et al: Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 2003; 72: 1117–1130.
Tutt A, Robson M, Garber JE et al: Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376: 235–244.
Audeh MW, Carmichael J, Penson RT et al: Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010; 376: 245–251.
Mann GJ, Thorne H, Balleine RL et al: Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res 2006; 8: R12.
Spiegelman JI, Mindrinos MN, Oefner PJ : High-accuracy DNA sequence variation screening by DHPLC. Biotechniques 2000; 29: 1084–1090, 1092.
Duponchel C, Di RC, Cicardi M, Tosi M : Rapid detection by fluorescent multiplex PCR of exon deletions and duplications in the C1 inhibitor gene of hereditary angioedema patients. Hum Mutat 2001; 17: 61–70.
Caux-Moncoutier V, Castera L, Tirapo C et al: EMMA, a cost- and time-effective diagnostic method for simultaneous detection of point mutations and large-scale genomic rearrangements: application to BRCA1 and BRCA2 in 1525 patients. Hum Mutat 2011; 32: 325–334.
Wittwer CT : High-resolution DNA melting analysis: advancements and limitations. Hum Mutat 2009; 30: 857–859.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G : Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002; 30: e57.
Walsh T, Casadei S, Coats KH et al: Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 2006; 295: 1379–1388.
Bubien V, Bonnet F, Brouste V et al: High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet 2013; 50: 255–263.
Giardiello FM, Brensinger JD, Tersmette AC et al: Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology 2000; 119: 1447–1453.
Pharoah PD, Guilford P, Caldas C : Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology 2001; 121: 1348–1353.
Meindl A, Hellebrand H, Wiek C et al: Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010; 42: 410–414.
Byrnes GB, Southey MC, Hopper JL : Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res 2008; 10: 208.
Rehm HL, Bale SJ, Bayrak-Toydemir P et al: ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013; 15: 733–747.
Walsh T, Lee MK, Casadei S et al: Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA 2010; 107: 12629–12633.
Craig DW, Pearson JV, Szelinger S et al: Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods 2008; 5: 887–893.
Kenny EM, Cormican P, Gilks WP et al: Multiplex target enrichment using DNA indexing for ultra-high throughput SNP detection. DNA Res 2011; 18: 31–38.
Xie C, Tammi MT : CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 2009; 10: 80.
Li H, Handsaker B, Wysoker A et al: The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Caputo S, Benboudjema L, Sinilnikova O, Rouleau E, Beroud C, Lidereau R : Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res 2012; 40: D992–1002.
Petitjean A, Mathe E, Kato S et al: Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007; 28: 622–629.
Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A : Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum Mutat 2008; 29: 1342–1354.
Houdayer C, Caux-Moncoutier V, Krieger S et al: Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat 2012; 33: 1228–1238.
Machado PM, Brandao RD, Cavaco BM et al: Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol 2007; 25: 2027–2034.
Tinat J, Bougeard G, Baert-Desurmont S et al: 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol 2009; 27: e108–e109.
Li J, Lupat R, Amarasinghe KC et al: CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012; 28: 1307–1313.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z : Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009; 25: 2865–2871.
Tarabeux J, Zeitouni B, Moncoutier V et al: Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet 2013, e-pub ahead of print 14 August 2013 doi:10.1038/ejhg.2013.181.
Masciari S, Dillon DA, Rath M et al: Breast cancer phenotype in women with TP53 germline mutations: a Li-Fraumeni syndrome consortium effort. Breast Cancer Res Treat 2012; 133: 1125–1130.
Gonzalez KD, Noltner KA, Buzin CH et al: Beyond Li Fraumeni syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol 2009; 27: 1250–1256.
Heymann S, Delaloge S, Rahal A et al: Radio-induced malignancies after breast cancer postoperative radiotherapy in patients with Li-Fraumeni syndrome. Radiat Oncol 2010; 5: 104.
Limacher JM, Frebourg T, Natarajan-Ame S, Bergerat JP : Two metachronous tumors in the radiotherapy fields of a patient with Li-Fraumeni syndrome. Int J Cancer 2001; 96: 238–242.
Ferrarini A, Auteri-Kaczmarek A, Pica A et al: Early occurrence of lung adenocarcinoma and breast cancer after radiotherapy of a chest wall sarcoma in a patient with a de novo germline mutation in TP53. Fam Cancer 2011; 10: 187–192.
Bonadona V, Bonaiti B, Olschwang S et al: Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011; 305: 2304–2310.
Lindor NM, Petersen GM, Hadley DW et al: Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA 2006; 296: 1507–1517.
Walsh T, Casadei S, Lee MK et al: Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA 2011; 108: 18032–18037.
Acknowledgements
We thank the high-throughput sequencing platform of Basse-Normandie SéSAME (Sequencing for Health, Agronomy, the Sea and the Environment) for technological support and the Institut National du Cancer (INCa) for funding. We are grateful to Mary-Claire King for providing the design of the first NGS panel and to Mario Tosi for critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Castéra, L., Krieger, S., Rousselin, A. et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet 22, 1305–1313 (2014). https://doi.org/10.1038/ejhg.2014.16
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.16
Keywords
This article is cited by
-
A comprehensive analysis of Fanconi anemia genes in Chinese patients with high-risk hereditary breast cancer
Journal of Cancer Research and Clinical Oncology (2023)
-
Germline variants associated with breast cancer in Khakass women of North Asia
Molecular Biology Reports (2023)
-
Feasibility of targeted cascade genetic testing in the family members of BRCA1/2 gene pathogenic variant/likely pathogenic variant carriers
Scientific Reports (2022)
-
Increased incidence of pathogenic variants in ATM in the context of testing for breast and ovarian cancer predisposition
Journal of Human Genetics (2022)
-
Identification of hereditary breast and ovarian cancer germline variants in Granada (Spain): NGS perspective
Molecular Genetics and Genomics (2022)