Abstract
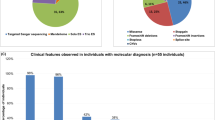
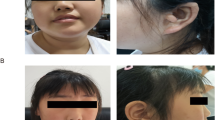
Intellectual disability (ID) has an estimated prevalence of 2–3%. Due to its extreme heterogeneity, the genetic basis of ID remains elusive in many cases. Recently, whole exome sequencing (WES) studies revealed that a large proportion of sporadic cases are caused by de novo gene variants. To identify further genes involved in ID, we performed WES in 250 patients with unexplained ID and their unaffected parents and included exomes of 51 previously sequenced child–parents trios in the analysis. Exome analysis revealed de novo intragenic variants in SET domain-containing 5 (SETD5) in two patients. One patient carried a nonsense variant, and the other an 81 bp deletion located across a splice-donor site. Chromosomal microarray diagnostics further identified four de novo non-recurrent microdeletions encompassing SETD5. CRISPR/Cas9 mutation modelling of the two intragenic variants demonstrated nonsense-mediated decay of the resulting transcripts, pointing to a loss-of-function (LoF) and haploinsufficiency as the common disease-causing mechanism of intragenic SETD5 sequence variants and SETD5-containing microdeletions. In silico domain prediction of SETD5, a predicted SET domain-containing histone methyltransferase (HMT), substantiated the presence of a SET domain and identified a novel putative PHD domain, strengthening a functional link to well-known histone-modifying ID genes. All six patients presented with ID and certain facial dysmorphisms, suggesting that SETD5 sequence variants contribute substantially to the microdeletion 3p25.3 phenotype. The present report of two SETD5 LoF variants in 301 patients demonstrates a prevalence of 0.7% and thus SETD5 variants as a relatively frequent cause of ID.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Flore LA, Milunsky JM : Updates in the genetic evaluation of the child with global developmental delay or intellectual disability. Semin Pediatr Neurol 2012; 19: 173–180.
Ropers HH : Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet 2010; 11: 161–187.
Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682.
de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929.
He X, Sanders SJ, Liu L et al: Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet 2013; 9: e1003671.
Sanders SJ, Murtha MT, Gupta AR et al: De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012; 485: 237–241.
Malmgren H, Sahlen S, Wide K, Lundvall M, Blennow E : Distal 3p deletion syndrome: detailed molecular cytogenetic and clinical characterization of three small distal deletions and review. Am J Med Genet A 2007; 143A: 2143–2149.
Shuib S, McMullan D, Rattenberry E et al: Microarray based analysis of 3p25-p26 deletions (3p- syndrome). Am J Med Genet A 2009; 149A: 2099–2105.
Peltekova IT, Macdonald A, Armour CM : Microdeletion on 3p25 in a patient with features of 3p deletion syndrome. Am J Med Genet A 2012; 158A: 2583–2586.
Kleefstra T, Brunner HG, Amiel J et al: Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet 2006; 79: 370–377.
Zweier M, Gregor A, Zweier C et al: Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat 2010; 31: 722–733.
Gunnarsson C, Foyn Bruun C : Molecular characterization and clinical features of a patient with an interstitial deletion of 3p25.3-p26.1. Am J Med Genet A 2010; 152A: 3110–3114.
Kellogg G, Sum J, Wallerstein R : Deletion of 3p25.3 in a patient with intellectual disability and dysmorphic features with further definition of a critical region. Am J Med Genet A 2013; 161A: 1405–1408.
Riess A, Grasshoff U, Schaferhoff K et al: Interstitial 3p25.3-p26.1 deletion in a patient with intellectual disability. Am J Med Genet A 2012; 158A: 2587–2590.
Pinto D, Delaby E, Merico D et al: Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 2014; 94: 677–694.
Grozeva D, Carss K, Spasic-Boskovic O et al: De novo loss-of-function mutations in setd5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet 2014; 94: 618–624.
Dokudovskaya S, Waharte F, Schlessinger A et al: A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 2011; 10: 006478.
van Dam TJ, Townsend MJ, Turk M et al: Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci USA 2013; 110: 6943–6948.
Cong L, Ran FA, Cox D et al: Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F : Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–2308.
Ottaviani D, Lecain M, Sheer D : The role of microhomology in genomic structural variation. Trends Genet 2014; 30: 85–94.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D : MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576.
Rea S, Eisenhaber F, O'Carroll D et al: Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406: 593–599.
Bienz M : The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 2006; 31: 35–40.
Jenuwein T, Laible G, Dorn R, Reuter G : SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 1998; 54: 80–93.
Sander JD, Joung JK : CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014; 32: 347–355.
Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB : Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013; 9: e1003709.
American Psychiatric Association AAP. Diagnostic and Statistical Manual of Mental Disorders 5th edn. Arlington, VA, USA: American Psychiatric Publishing, 2013.
van Bokhoven H : Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet 2011; 45: 81–104.
Tan M, Luo H, Lee S et al: Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011; 146: 1016–1028.
Kleefstra T, Schenck A, Kramer JM, van Bokhoven H : The genetics of cognitive epigenetics. Neuropharmacology 2014; 80: 83–89.
Martin C, Zhang Y : The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6: 838–849.
Acknowledgements
We are grateful to the patients and their families for participating in this study. We thank the ‘German Mental Retardation Network’ (MRNET) and Sabine Kaya, Daniela Falkenstein and Mike Liu for their excellent technical assistance. This work was supported in part by the German Ministry of Research and Education (grant numbers 01GS08164, 01GS08167, 01GS08163, German Mental Retardation Network) as part of the National Genome Research Network and by the European Commission’s FP7 CHERISH programme (grant number 223692).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Kuechler, A., Zink, A., Wieland, T. et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur J Hum Genet 23, 753–760 (2015). https://doi.org/10.1038/ejhg.2014.165
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.165
This article is cited by
-
SETD5 haploinsufficiency affects mitochondrial compartment in neural cells
Molecular Autism (2023)
-
SETD5 modulates homeostasis of hematopoietic stem cells by mediating RNA Polymerase II pausing in cooperation with HCF-1
Leukemia (2022)
-
Spatiotemporal dynamics of SETD5-containing NCoR–HDAC3 complex determines enhancer activation for adipogenesis
Nature Communications (2021)
-
SET domain containing protein 5 (SETD5) enhances tumor cell invasion and is associated with a poor prognosis in non-small cell lung cancer patients
BMC Cancer (2019)
-
Setd5 haploinsufficiency alters neuronal network connectivity and leads to autistic-like behaviors in mice
Translational Psychiatry (2019)