Abstract
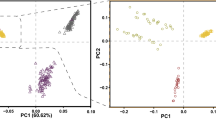
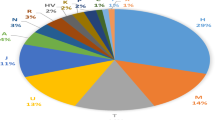
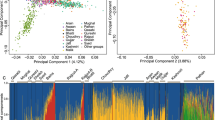
Druze individuals rarely marry outside their faith (often practicing consanguinity) and are thus believed to form a genetic isolate. To comprehensively characterize the genetic structure of the Druze population, we recruited and genotyped 40 parent-offspring trios from the Upper Galilee in Israel and the Golan Heights, attempting to capture different extended families (clans) across various geographical locations. Principal component (PC) and ADMIXTURE analyses demonstrated that Druze are close to, yet distinct from, other Middle-Eastern groups (Bedouins and Palestinians), supporting the Druze’s Middle-Eastern origin and their recent genetic isolation. Reconstruction of the Druze demographic history using identical-by-descent (IBD) segments suggested an ≈15-fold reduction in population size taking place ≈22–47 generations ago, close to the documented time of the foundation of the Druze faith at the 11th century. Combining the Galilee and Golan Druze genotypes with previously published data on Druze from the Carmel (Israel) and Lebanon demonstrated that all four Druze communities are genetically distinct. The Lebanese group shared less IBD segments (within the group and with other groups) compared with the Israeli Druze and showed higher heterozygosity (suggesting less consanguinity), but was less diverse in PC space. These findings suggest complex recent and ancient demographic history of the Druze population.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Hitti PK : Origins of the Druze People and Religion: with Extracts from Their Sacred Writings, Columbia University Oriental Studies 28 (new ed.). London: Saqi, 2007 [1924], pp 13–14
Swayd S : The Druzes: An Annotated Bibliography. Kirkland, WA, USA: ISES Publications, 1998.
Cohen N, Czamanski D, Hefetz A : Internal migration of Ethno-national Minorities: the case of Arabs in Israel. Int Migr 2012, e-pub ahead of print 29 November 2012 doi:10.1111/imig.12014
Hitti PK : The Origins of the Druze People and Religion. Netlancers Inc, 2014
Vardi-Saliternik R, Friedlander Y, Cohen T : Consanguinity in a population sample of Israeli Muslim Arabs, Christian Arabs and Druze. Ann Hum Biol 2002; 29: 422–431.
Fares F, Axelord Ran S, David M et al: Identification of two mutations for ataxia telangiectasia among the Druze community. Prenat Diagn 2004; 24: 358–362.
Filon D, Oron V, Krichevski S et al: Diversity of beta-globin mutations in Israeli ethnic groups reflects recent historic events. Am J Hum Genet 1994; 54: 836–843.
Landsberger D, Meiner V, Reshef A et al: A nonsense mutation in the LDL receptor gene leads to familial hypercholesterolemia in the Druze sect. Am J Hum Genet 1992; 50: 427–433.
Ronen O, Cohen SB, Rund D : Evaluating frequencies of thiopurine S-methyl transferase (TPMT) variant alleles in Israeli ethnic subpopulations using DNA analysis. Isr Med Assoc J 2010; 12: 721–725.
Karban A, Krivoy N, Elkin H et al: Non-Jewish Israeli IBD patients have significantly higher glutathione S-transferase GSTT1-null frequency. Dig Dis Sci 2011; 56: 2081–2087.
Efrati E, Elkin H, Sprecher E, Krivoy N : Distribution of CYP2C9 and VKORC1 risk alleles for warfarin sensitivity and resistance in the Israeli population. Curr Drug Saf 2010; 5: 190–193.
Klein P, Weinberger A, Altmann VJ, Halabi S, Fachereldeen S, Krause I : Prevalence of Behcet's disease among adult patients consulting three major clinics in a Druze town in Israel. Clin Rheumatol 2010; 29: 1163–1166.
Shlush LI, Behar DM, Yudkovsky G et al: The Druze: a population genetic refugium of the Near East. PLoS One 2008; 3: e2105
Zalloua PA, Xue Y, Khalife J et al: Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet 2008; 82: 873–882.
Li JZ, Absher DM, Tang H et al: Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104.
Behar DM, Yunusbayev B, Metspalu M et al: The genome-wide structure of the Jewish people. Nature 2010; 466: 238–242.
Haber M, Gauguier D, Youhanna S et al: Genome-wide diversity in the levant reveals recent structuring by culture. PLoS Genet 2013; 9: e1003316.
Moorjani P, Patterson N, Hirschhorn JN et al: The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet 2011; 7: e1001373.
Cann HM, de Toma C, Cazes L et al: A human genome diversity cell line panel. Science 2002; 296: 261–262
Rosenberg NA : Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet 2006; 70: 841–847.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575
Delaneau O, Marchini J, Zagury JF : A linear complexity phasing method for thousands of genomes. Nat Methods 2012; 9: 179–181.
Alexander DH, Novembre J, Lange K : Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19: 1655–1664.
International HapMap Consortium, Frazer KA, Ballinger DG et al: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861
Gusev A, Lowe JK, Stoffel M et al: Whole population, genome-wide mapping of hidden relatedness. Genome Res 2009; 19: 318–326.
Durand EY, Eriksson N, McLean CY : Reducing pervasive false positive identical-by-descent segments detected by large-scale pedigree analysis. Mol Biol Evol 2014; 31: 2212–2222.
Meyer LR, Zweig AS, Hinrichs AS et al: The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 2013; 41: D64–D69
Gusev A, Palamara PF, Aponte G et al: The architecture of long-range haplotypes shared within and across populations. Mol Biol Evol 2012; 29: 473–486.
Palamara PF, Lencz T, Darvasi A, Pe'er I : Length distributions of identity by descent reveal fine-scale demographic history. Am J Hum Genet 2012; 91: 809–822.
Carmi S, Palamara PF, Vacic V, Lencz T, Darvasi A, Pe'er I : The variance of identity-by-descent sharing in the Wright-Fisher Model. Genetics 2013; 193: 911–928.
Wakeley J : Coalescent Theory: An Introduction. Greenwood Village, Colorado, USA: Roberts & Company Publishers, 2009.
Browning SR, Browning BL : Identity by descent between distant relatives: detection and applications. Annu Rev Genet 2012; 46: 617–633.
Ralph P, Coop G : The geography of recent genetic ancestry across Europe. PLoS Biol 2013; 11: e1001555.
Atzmon G, Hao L, Pe'er I et al: Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet 2010; 86: 850–859.
Magalhaes TR, Casey JP, Conroy J et al: HGDP and HapMap analysis by Ancestry Mapper reveals local and global population relationships. PLoS One 2012; 7: e49438
Kopelman NM, Stone L, Wang C et al: Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet 2009; 10: 80.
Huang L, Li Y, Singleton AB et al: Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet 2009; 84: 235–250.
Hodoglugil U, Mahley RW : Turkish population structure and genetic ancestry reveal relatedness among Eurasian populations. Ann Hum Genet 2012; 76: 128–141.
Scally A, Durbin R : Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet 2012; 13: 745–753.
McVean G : A genealogical interpretation of principal components analysis. PLoS Genet 2009; 5: e1000686.
Acknowledgements
This study was in part sponsored by a grant from the Binational Science Foundation (BSF) to EF and Harry Ostrer and generous donations from private donors to the Golan for Development organization and the Oncology Institute at the Ziv medical center. SC acknowledges financial support from the Human Frontier Science Program and GA NIH (1R01AG042188).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Zidan, J., Ben-Avraham, D., Carmi, S. et al. Genotyping of geographically diverse Druze trios reveals substructure and a recent bottleneck. Eur J Hum Genet 23, 1093–1099 (2015). https://doi.org/10.1038/ejhg.2014.218
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.218
This article is cited by
-
Genetic Basis of Delayed Hypersensitivity Reactions to Drugs in Jewish and Arab Populations
Pharmaceutical Research (2018)
-
Genomic insights into the population structure and history of the Irish Travellers
Scientific Reports (2017)
-
The Israeli national population program of genetic carrier screening for reproductive purposes
Genetics in Medicine (2016)