Abstract
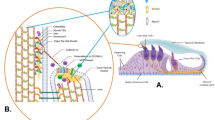
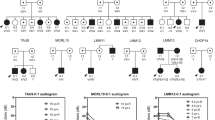
In a consanguineous Turkish family diagnosed with autosomal recessive nonsyndromic hearing impairment (arNSHI), a homozygous region of 47.4 Mb was shared by the two affected siblings on chromosome 6p21.1-q15. This region contains 247 genes including the known deafness gene MYO6. No pathogenic variants were found in MYO6, neither with sequence analysis of the coding region and splice sites nor with mRNA analysis. Subsequent candidate gene evaluation revealed CLIC5 as an excellent candidate gene. The orthologous mouse gene is mutated in the jitterbug mutant that exhibits progressive hearing impairment and vestibular dysfunction. Mutation analysis of CLIC5 revealed a homozygous nonsense mutation c.96T>A (p.(Cys32Ter)) that segregated with the hearing loss. Further analysis of CLIC5 in 213 arNSHI patients from mostly Dutch and Spanish origin did not reveal any additional pathogenic variants. CLIC5 mutations are thus not a common cause of arNSHI in these populations. The hearing loss in the present family had an onset in early childhood and progressed from mild to severe or even profound before the second decade. Impaired hearing is accompanied by vestibular areflexia and in one of the patients with mild renal dysfunction. Although we demonstrate that CLIC5 is expressed in many other human tissues, no additional symptoms were observed in these patients. In conclusion, our results show that CLIC5 is a novel arNSHI gene involved in progressive hearing impairment, vestibular and possibly mild renal dysfunction in a family of Turkish origin.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Mehl AL, Thomson V : The Colorado newborn hearing screening project, 1992-1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 2002; 109: E7.
Dror AA, Avraham KB : Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet 2009; 43: 411–437.
Schraders M, Lee K, Oostrik J et al: Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet 2010; 86: 138–147.
Shearer AE, Smith RJ : Genetics: advances in genetic testing for deafness. Curr Opin Pediatr 2012; 24: 679–686.
Delmaghani S, Aghaie A, Michalski N, Bonnet C, Weil D, Petit C : Defect in the gene encoding the EAR/EPTP domain-containing protein TSPEAR causes DFNB98 profound deafness. Hum Mol Genet 2012; 21: 3835–3844.
Sirmaci A, Erbek S, Price J et al: A truncating mutation in SERPINB6 is associated with autosomal-recessive nonsyndromic sensorineural hearing loss. Am J Hum Genet 2010; 86: 797–804.
von Ameln S, Wang G, Boulouiz R et al: A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am J Hum Genet 2012; 91: 919–927.
Friedman LM, Dror AA, Avraham KB : Mouse models to study inner ear development and hereditary hearing loss. Int J Dev Biol 2007; 51: 609–631.
Gagnon LH, Longo-Guess CM, Berryman M et al: The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci 2006; 26: 10188–10198.
Theunissen EJ, Huygen PL, Folgering HT : Vestibular hyperreactivity and hyperventilation. Clin Otolaryngol Allied Sci 1986; 11: 161–169.
Livak KJ, Schmittgen TD : Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408.
Luijendijk MW, van de Pol TJ, van Duijnhoven G et al: Cloning, characterization, and mRNA expression analysis of novel human fetal cochlear cDNAs. Genomics 2003; 82: 480–490.
Pfaffl MW : A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Bosman AJ SG : Woordenlijst Voor Spraakaudiometrie (Compact Disc) Gouda. Electro Medical Instruments bv & Veenhuis Medical Audio bv: The Netherlands, 1992.
Prolo LM, Vogel H, Reimer RJ : The lysosomal sialic acid transporter sialin is required for normal CNS myelination. J Neurosci 2009; 29: 15355–15365.
Nagy E, Maquat LE : A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998; 23: 198–199.
Pierchala BA, Munoz MR, Tsui CC : Proteomic analysis of the slit diaphragm complex: CLIC5 is a protein critical for podocyte morphology and function. Kidney Int 2010; 78: 868–882.
Wegner B, Al-Momany A, Kulak SC et al: CLIC5A, a component of the ezrin-podocalyxin complex in glomeruli, is a determinant of podocyte integrity. Am J Physiol Renal Physiol 2010; 298: F1492–F1503.
Bradford EM, Miller ML, Prasad V et al: CLIC5 mutant mice are resistant to diet-induced obesity and exhibit gastric hemorrhaging and increased susceptibility to torpor. Am J Physiol Regul Integr Comp Physiol 2010; 298: R1531–R1542.
Dulhunty A, Gage P, Curtis S, Chelvanayagam G, Board P : The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J Biol Chem 2001; 276: 3319–3323.
Ashley RH : Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins (Review). Mol Membr Biol 2003; 20: 1–11.
Berryman M, Bretscher A : Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli. Mol Biol Cell 2000; 11: 1509–1521.
Jiang L, Phang JM, Yu J et al: CLIC proteins, ezrin, radixin, moesin and the coupling of membranes to the actin cytoskeleton: a smoking gun? Biochim Biophys Acta 2014; 1838: 643–657.
Salles FT, Andrade LR, Tanda S et al: CLIC5 stabilizes membrane-actin filament linkages at the base of hair cell stereocilia in a molecular complex with radixin, taperin, and myosin VI. Cytoskeleton (Hoboken) 2013; 71: 61–78.
Sakaguchi H, Tokita J, Naoz M, Bowen-Pope D, Gov NS, Kachar B : Dynamic compartmentalization of protein tyrosine phosphatase receptor Q at the proximal end of stereocilia: implication of myosin VI-based transport. Cell Motil Cytoskeleton 2008; 65: 528–538.
Kitajiri S, Fukumoto K, Hata M et al: Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol 2004; 166: 559–570.
Goodyear RJ, Legan PK, Wright MB et al: A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci 2003; 23: 9208–9219.
Self T, Sobe T, Copeland NG, Jenkins NA, Avraham KB, Steel KP : Role of myosin VI in the differentiation of cochlear hair cells. Dev Biol 1999; 214: 331–341.
Acknowledgements
We are grateful to the family for their participation in this study. We thank Saskia van der Velde-Visser, Marloes Steehouwer and Irene Janssen for excellent technical assistance, Ersan Kalay for providing Turkish control DNA samples, Arjan de Brouwer for his contribution to the NMD analysis and Rob Collin for SNP-array analysis. This work was financially supported by grants from the Heinsius Houbolt Foundation (to HK), The Oticon Foundation (09-3742, to HK), ZorgOnderzoek Nederland/Medische Wetenschappen (40-00812-98-09047, to HK, 90700388 to RJEP and 016.136.088 to MS), the Netherlands Genomics Initiative (40-41009-98-9073, to MS) and Instituto de Salud Carlos III (FIS PI11/00612, to IdC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Seco, C., Oonk, A., Domínguez-Ruiz, M. et al. Progressive hearing loss and vestibular dysfunction caused by a homozygous nonsense mutation in CLIC5. Eur J Hum Genet 23, 189–194 (2015). https://doi.org/10.1038/ejhg.2014.83
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.83
This article is cited by
-
ARNSHL gene identification: past, present and future
Molecular Genetics and Genomics (2022)
-
Identification of autosomal recessive nonsyndromic hearing impairment genes through the study of consanguineous and non-consanguineous families: past, present, and future
Human Genetics (2022)
-
Inherent flexibility of CLIC6 revealed by crystallographic and solution studies
Scientific Reports (2018)
-
Genetic contribution to vestibular diseases
Journal of Neurology (2018)
-
The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands
European Journal of Human Genetics (2017)