Abstract
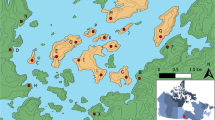
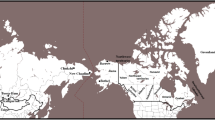
The peopling of Greenland has a complex history shaped by population migrations, isolation and genetic drift. The Greenlanders present a genetic heritage with components of European and Inuit groups; previous studies using uniparentally inherited markers in Greenlanders have reported evidence of a sex-biased, admixed genetic background. This work further explores the genetics of the Greenlanders by analysing autosomal and X-chromosomal data to obtain deeper insights into the factors that shaped the genetic diversity in Greenlanders. Fourteen Greenlandic subsamples from multiple geographical settlements were compared to assess the level of genetic substructure in the Greenlandic population. The results showed low levels of genetic diversity in all sets of the genetic markers studied, together with an increased number of X-chromosomal loci in linkage disequilibrium in relation to the Danish population. In the broader context of worldwide populations, Greenlanders are remarkably different from most populations, but they are genetically closer to some Inuit groups from Alaska. Admixture analyses identified an Inuit component in the Greenlandic population of approximately 80%. The sub-populations of Ammassalik and Nanortalik are the least diverse, presenting the lowest levels of European admixture. Isolation-by-distance analyses showed that only 16% of the genetic substructure of Greenlanders is most likely to be explained by geographic barriers. We suggest that genetic drift and a differentiated settlement history around the island explain most of the genetic substructure of the population in Greenland.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Gilbert MT, Kivisild T, Gronnow B et al: Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science 2008; 320: 1787–1789.
McGhee R : Radiocarbon dating and the timing of the Thule migration; in Applet M, Berglund J, Gullov HC, (eds): Identities and Cultural Contacts in the Arctic. Copenhagen, Denmark: Danish National Museum & Danish Polar Center, 2000, pp 181–191.
Morrison D : The arrival of the Inuit: Amundsen Gulf and the Thule migration; in Applet M, Berglund J, Gullov HC, (eds): Identities and Cultural Contacts in the Arctic. Copenhagen, Denmark: Danish National Museum & Danish Polar Center, 2000, pp 221–228.
Helgason A, Palsson G, Pedersen HS et al: mtDNA variation in Inuit populations of Greenland and Canada: migration history and population structure. Am J Phys Anthropol 2006; 130: 123–134.
Kleivan I : History of Norse Greenland; in Damas D, (ed): Handbook of North American Indians, Vol 5. The Arctic. Washington DC, USA: Smithsonian Institution, 1984; 549–555.
Bosch E, Calafell F, Rosser ZH et al: High level of male-biased Scandinavian admixture in Greenlandic Inuit shown by Y-chromosomal analysis. Hum Genet 2003; 112: 353–363.
Bosch E, Rosser ZH, Norby S et al: Y-chromosomal STR haplotypes in Inuit and Danish population samples. Forensic Sci Int 2003; 132: 228–232.
Sanchez JJ, Børsting C, Hernandez A et al: Y chromosome SNP haplogroups in Danes, Greenlanders and Somalis. Int Congr Ser 2004; 1261: 347–349.
Hallenberg C, Tomas C, Simonsen B et al: Y-chromosome STR haplotypes in males from Greenland. Forensic Sci Int Genet 2009; 3: e145–e146.
Saillard J, Forster P, Lynnerup N et al: MtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 2000; 67: 718–726.
Rasmussen M, Li Y, Lindgreen S et al: Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 2010; 463: 757–762.
Schaffner SF : The X chromosome in population genetics. Nat Rev Genet 2004; 5: 43–51.
Pereira R, Pereira V, Gomes I et al: A method for the analysis of 32 X chromosome insertion deletion polymorphisms in a single PCR. Int J Legal Med 2012; 126: 97–105.
Tomas C, Sanchez JJ, Barbaro A et al: X-chromosome SNP analyses in 11 human Mediterranean populations show a high overall genetic homogeneity except in North-west Africans (Moroccans). BMC Evol Biol 2008; 8: 75.
Excoffier L, Lischer HE : Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010; 10: 564–567.
Raymond M, Rousset F : An exact test for population differentiation. Evolution 1995; 49: 1280–1283.
Takezaki N, Nei M, Tamura K : POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 2010; 27: 747–752.
Holm S : A simple sequentially rejective multiple test procedure. Scand J Stat 1979; 6: 65–70.
Bohonak AJ : IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 2002; 93: 153–154.
Rousset F : Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997; 145: 1219–1228.
Pritchard JK, Stephens M, Donnelly P : Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–959.
Jakobsson M, Rosenberg NA : CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007; 23: 1801–1806.
Rosenberg NA : Distruct: a program for the graphical display of population structure. Mol Ecol Notes 2004; 4: 137–138.
Tomas C, Pereira V, Morling N : Analysis of 12 X-STRs in Greenlanders, Danes and Somalis using Argus X-12. Int J Legal Med 2012; 126: 121–128.
Jobling MA, Hurles M, Tyler-Smith C : Human Evolutionary Genetics: Origins, Peoples and Disease Kentucky, USA 2003.
Budowle B, Chidambaram A, Strickland L et al: Population studies on three Native Alaska population groups using STR loci. Forensic Sci Int 2002; 129: 51–57.
Jensen JL, Bohonak AJ, Kelley ST : Isolation by distance, web service. BMC Genet 2005; 6: 13.
Pereira V, Tomas C, Sanchez JJ et al: Study of 25 X-chromosome single nucleotide polymorphisms in African and Asian populations. Forensic Sci Int Gen Suppl Ser 2011; 3: e139–e140.
Acknowledgements
We thank all the personnel at the Department of Forensic Medicine in Copenhagen who were involved in the sample collection and typing. We also thank the Unidade de Xenética, Instituto de Ciencias Forenses and Departamento de Anatomía Patolóxica e Ciencias Forenses, Universidade de Santiago de Compostela for providing samples from Taiwan. This work was partially supported by Ellen and Aage Andersen’s Foundation. IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by FCT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Pereira, V., Tomas, C., Sanchez, J. et al. The peopling of Greenland: further insights from the analysis of genetic diversity using autosomal and X-chromosomal markers. Eur J Hum Genet 23, 245–251 (2015). https://doi.org/10.1038/ejhg.2014.90
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.90
This article is cited by
-
Genetic characterization of 32 X-InDels in a population sample from São Paulo State (Brazil)
International Journal of Legal Medicine (2019)
-
A racial classification for medical genetics
Philosophical Studies (2018)
-
Determination of NAT2 acetylation status in the Greenlandic population
Archives of Toxicology (2016)