Abstract
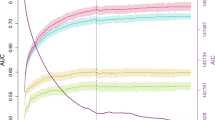
The global demand for products that effectively prevent the development of male-pattern baldness (MPB) has drastically increased. However, there is currently no established genetic model for the estimation of MPB risk. We conducted a prediction analysis using single-nucleotide polymorphisms (SNPs) identified from previous GWASs of MPB in a total of 2725 German and Dutch males. A logistic regression model considering the genotypes of 25 SNPs from 12 genomic loci demonstrates that early-onset MPB risk is predictable at an accuracy level of 0.74 when 14 SNPs were included in the model, and measured using the area under the receiver-operating characteristic curves (AUC). Considering age as an additional predictor, the model can predict normal MPB status in middle-aged and elderly individuals at a slightly lower accuracy (AUC 0.69–0.71) when 6–11 SNPs were used. A variance partitioning analysis suggests that 55.8% of early-onset MPB genetic liability can be explained by common autosomal SNPs and 23.3% by X-chromosome SNPs. For normal MPB status in elderly individuals, the proportion of explainable variance is lower (42.4% for autosomal and 9.8% for X-chromosome SNPs). The gap between GWAS findings and the variance partitioning results could be explained by a large body of common DNA variants with small effects that will likely be identified in GWAS of increased sample sizes. Although the accuracy obtained here has not reached a clinically desired level, our model was highly informative for up to 19% of Europeans, thus may assist decision making on early MPB intervention actions and in forensic investigations.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Norwood OT : Male pattern baldness: classification and incidence. South Med J 1975; 68: 1359–1365.
Nyholt DR, Gillespie NA, Heath AC, Martin NG : Genetic basis of male pattern baldness. J Invest Dermatol 2003; 121: 1561–1564.
Hamilton JB : Patterned loss of hair in man; types and incidence. Ann N Y Acad Sci 1951; 53: 708–728.
Cash TF : The psychological effects of androgenetic alopecia in men. J Am Acad Dermatol 1992; 26: 926–931.
Sinclair R : Male pattern androgenetic alopecia. BMJ 1998; 317: 865–869.
Kayser M, de Knijff P : Improving human forensics through advances in genetics, genomics and molecular biology. Nat Rev Genet 2011; 12: 179–192.
Kayser M : Forensic DNA phenotyping: predicting human appearance from crime scene material for investigative purposes. Forensic Sci Int Genet 2015; 18: 33–48.
Zubakov D, Liu F, van Zelm MC et al: Estimating human age from T-cell DNA rearrangements. Curr Biol 2010; 20: R970–R971.
Zbiec-Piekarska R, Spolnicka M, Kupiec T et al: Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci Int Genet 2015; 14: 161–167.
Rexbye H, Petersen I, Iachina M et al: Hair loss among elderly men: etiology and impact on perceived age. J Gerontol A Biol Sci Med Sci 2005; 60: 1077–1082.
Ellis JA, Stebbing M, Harrap SB : Polymorphism of the androgen receptor gene is associated with male pattern baldness. J Invest Dermatol 2001; 116: 452–455.
Prodi DA, Pirastu N, Maninchedda G et al: EDA2R is associated with androgenetic alopecia. J Invest Dermatol 2008; 128: 2268–2270.
Brockschmidt FF, Heilmann S, Ellis JA et al: Susceptibility variants on chromosome 7p21.1 suggest HDAC9 as a new candidate gene for male-pattern baldness. Br J Dermatol 2011; 165: 1293–1302.
Hillmer AM, Brockschmidt FF, Hanneken S et al: Susceptibility variants for male-pattern baldness on chromosome 20p11. Nat Genet 2008; 40: 1279–1281.
Richards JB, Yuan X, Geller F et al: Male-pattern baldness susceptibility locus at 20p11. Nat Genet 2008; 40: 1282–1284.
Li R, Brockschmidt FF, Kiefer AK et al: Six novel susceptibility loci for early-onset androgenetic alopecia and their unexpected association with common diseases. PLoS Genet 2012; 8: e1002746.
Heilmann S, Kiefer AK, Fricker N et al: Androgenetic alopecia: identification of four genetic risk loci and evidence for the contribution of WNT signaling to its etiology. J Invest Dermatol 2013; 133: 1489–1496.
Hofman A, Darwish Murad S, van Duijn CM et al: The Rotterdam Study: 2014 objectives and design update. Eur J Epidemiol 2013; 28: 889–926.
Taylor R, Matassa J, Leavy JE, Fritschi L : Validity of self reported male balding patterns in epidemiological studies. BMC Public Health 2004; 4: 60.
Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS : The effect of genetic drift in a young genetically isolated population. Ann Hum Genet 2005; 69: 288–295.
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR : MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834.
Marchini J, Howie B, Myers S, McVean G, Donnelly P : A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913.
Akaike H : Citation classic—a new look at the statistical-model identification. CC/Eng Technol Appl Sci 1981; 51: 22–22.
Zweig MH, Campbell G : Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 1993; 39: 561–577.
Yang J, Lee SH, Goddard ME, Visscher PM : GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ : Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7.
Patterson N, Price AL, Reich D : Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Visser M, Palstra RJ, Kayser M : Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum Mol Genet 2014; 23: 5750–5762.
Visser M, Kayser M, Palstra RJ : HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res 2012; 22: 446–455.
Lee WS, Lee HJ : Characteristics of androgenetic alopecia in asian. Ann Dermatol 2012; 24: 243–252.
Hoffmann R : Male androgenetic alopecia. Clin Exp Dermatol 2002; 27: 373–382.
Birch MP, Messenger AG : Genetic factors predispose to balding and non-balding in men. Eur J Dermatol 2001; 11: 309–314.
Liu F, Wollstein A, Hysi PG et al: Digital quantification of human eye color highlights genetic association of three new loci. PLoS Genet 2010; 6: e1000934.
Liu F, van Duijn K, Vingerling JR et al: Eye color and the prediction of complex phenotypes from genotypes. Curr Biol 2009; 19: R192–R193.
Liu F, Hendriks AEJ, Ralf A et al: Common DNA variants predict tall stature in Europeans. Hum Genet 2013; 133: 587–597.
Wood AR, Esko T, Yang J et al: Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014; 46: 1173–1186.
Acknowledgements
We thank Dr David Gunn for his valuable discussions and useful comments on the manuscript. We thank Pascal Arp; Mila Jhamai; Marijn Verkerk; Lizbeth Herrera; Marjolein Peters, MSc; Carolina Medina-Gomez, MSc; and Fernando Rivadeneira, MD PhD, for their help in creating the GWAS database, and Karol Estrada, PhD; Yurii Aulchenko, PhD; and Carolina Medina-Gomez, MSc, for the creation and analysis of imputed data. We thank Sophie Flohil, Emmilia Dowlatshahi, Robert van der Leest, Leonie Jacobs, Joris Verkouteren, Ella van der Voort, and Shmaila Talib for collecting the phenotype data in the RS. This work was supported in part by the Erasmus MC University Medical Center Rotterdam and funds from the Netherlands Genomics Initiative/Netherlands Organization of Scientific Research (NWO) within the framework of the Netherlands Consortium of Healthy Ageing (NCHA).
The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The GWAS data sets are supported by the Netherlands Organisation of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO), and the Netherlands Consortium for Healthy Aging (NCHA), project no. 050-060-810. FL is supported by Chinese Thousand Talent Program for Distinguished Young Scholars and MAH is supported by Unilever. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam; Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
ERF Study as a part of EUROSPAN (European Special Populations Research Network) was supported by the European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme ‘Quality of Life and Management of the Living Resources’ of 5th Framework Programme (no. QLG2-CT-2002-01254) as well as the FP7 project EUROHEADPAIN (no. 602633). High-throughput analysis of the ERF data was supported by joint grant from Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043).
The BONN Study is supported by Heinz Nixdorf Foundation, the German Ministry of Education and Science and the German Research Council (D Glass; Project SI 236/8-1, SI236/9-1, ER 155/6-1); German Research Council (D Glass; FOR 423); and the Life and Brain GmbH (Bonn, Germany; project grant). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Liu, F., Hamer, M., Heilmann, S. et al. Prediction of male-pattern baldness from genotypes. Eur J Hum Genet 24, 895–902 (2016). https://doi.org/10.1038/ejhg.2015.220
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2015.220
This article is cited by
-
Platelet-Rich Plasma for Treatment of Hair Loss Improves Patient-Reported Quality of Life
Aesthetic Plastic Surgery (2023)
-
Biology and Criminology: Data Practices and the Creation of Anatomic and Genomic Body ‘Types’
Critical Criminology (2023)
-
Exploring the possibility of predicting human head hair greying from DNA using whole-exome and targeted NGS data
BMC Genomics (2020)
-
DNA makes an appearance
Nature Biotechnology (2018)
-
Early-onset baldness and the risk of aggressive prostate cancer: findings from a case–control study
Cancer Causes & Control (2018)