Abstract
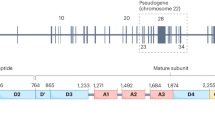
Low von Willebrand factor (VWF) levels are associated with bleeding symptoms and are a diagnostic criterion for von Willebrand disease, the most common inherited bleeding disorder. To date, it is unclear which genetic loci are associated with reduced VWF levels. Therefore, we conducted a meta-analysis of genome-wide association studies to identify genetic loci associated with low VWF levels. For this meta-analysis, we included 31 149 participants of European ancestry from 11 community-based studies. From all participants, VWF antigen (VWF:Ag) measurements and genome-wide single-nucleotide polymorphism (SNP) scans were available. Each study conducted analyses using logistic regression of SNPs on dichotomized VWF:Ag measures (lowest 5% for blood group O and non-O) with an additive genetic model adjusted for age and sex. An inverse-variance weighted meta-analysis was performed for VWF:Ag levels. A total of 97 SNPs exceeded the genome-wide significance threshold of 5 × 10−8 and comprised five loci on four different chromosomes: 6q24 (smallest P-value 5.8 × 10−10), 9q34 (2.4 × 10−64), 12p13 (5.3 × 10−22), 12q23 (1.2 × 10−8) and 13q13 (2.6 × 10−8). All loci were within or close to genes, including STXBP5 (Syntaxin Binding Protein 5) (6q24), STAB5 (stabilin-5) (12q23), ABO (9q34), VWF (12p13) and UFM1 (ubiquitin-fold modifier 1) (13q13). Of these, UFM1 has not been previously associated with VWF:Ag levels. Four genes that were previously associated with VWF levels (VWF, ABO, STXBP5 and STAB2) were also associated with low VWF levels, and, in addition, we identified a new gene, UFM1, that is associated with low VWF levels. These findings point to novel mechanisms for the occurrence of low VWF levels.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Ruggeri ZM : Old concepts and new developments in the study of platelet aggregation. J Clin Invest 2000; 105: 699–701.
Ruggeri ZM, Ware J : von Willebrand factor. FASEB J 1993; 7: 308–316.
Nichols WL, Hultin MB, James AH et al: von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA). Haemophilia 2008; 14: 171–232.
Bladbjerg EM, de Maat MP, Christensen K, Bathum L, Jespersen J, Hjelmborg J : Genetic influence on thrombotic risk markers in the elderly – a Danish twin study. J Thromb Haemost 2006; 4: 599–607.
Martinelli I : von Willebrand factor and factor VIII as risk factors for arterial and venous thrombosis. Semin Hematol 2005; 42: 49–55.
Koster T, Blann AD, Briet E, Vandenbroucke JP, Rosendaal FR : Role of clotting factor VIII in effect of von Willebrand factor on occurrence of deep-vein thrombosis. Lancet 1995; 345: 152–155.
Spiel AO, Gilbert JC, Jilma B : von Willebrand factor in cardiovascular disease: focus on acute coronary syndromes. Circulation 2008; 117: 1449–1459.
Smith A, Patterson C, Yarnell J, Rumley A, Ben-Shlomo Y, Lowe G : Which hemostatic markers add to the predictive value of conventional risk factors for coronary heart disease and ischemic stroke? The Caerphilly Study. Circulation 2005; 112: 3080–3087.
Sadler JE, Budde U, Eikenboom JC et al: Update on the pathophysiology and classification of von Willebrand disease: a report of the Subcommittee on von Willebrand Factor. J Thromb Haemost 2006; 4: 2103–2114.
Goodeve A, Eikenboom J, Castaman G et al: Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD). Blood 2007; 109: 112–121.
James PD, Notley C, Hegadorn C et al: The mutational spectrum of type 1 von Willebrand disease: results from a Canadian cohort study. Blood 2007; 109: 145–154.
Sadler JE : Low von Willebrand factor: sometimes a risk factor and sometimes a disease. Hematol Am Soc Hematol Educ Program 2009; 1: 106–112.
Sanders YV, Van der Bom JG, Isaacs A et al: CLEC4M and STXBP5 gene variations contribute to von Willebrand factor level variation in von Willebrand disease. J Thromb Haemost 2015; 13: 956–966.
Psaty BM, O'Donnell CJ, Gudnason V et al: Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009; 2: 73–80.
Hofman A, Breteler MM, van Duijn CM et al: The Rotterdam Study: objectives and design update. Eur J Epidemiol 2007; 22: 819–829.
The ARIC Investigators.: The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol 1989; 129: 687–702.
Dawber TR, Meadors GF, Moore FE Jr : Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health 1951; 41: 279–281.
Strachan DP, Rudnicka AR, Power C et al: Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int J Epidemiol 2007; 36: 522–531.
McQuillan R, Leutenegger AL, Abdel-Rahman R et al: Runs of homozygosity in European populations. Am J Hum Genet 2008; 83: 359–372.
Yang Q, Kathiresan S, Lin JP, Tofler GH, O'Donnell CJ : Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med Genet 2007; 8 (Suppl 1): S12.
Houlihan LM, Davies G, Tenesa A et al: Common variants of large effect in F12, KNG1, and HRG are associated with activated partial thromboplastin time. Am J Hum Genet 2010; 86: 626–631.
Rudan I, Campbell H, Rudan P : Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Coll Antropol 1999; 23: 531–546.
Hillege HL, Fidler V, Diercks GF et al: Urinary albumin excretion predicts cardiovascular and noncardiovascular mortality in general population. Circulation 2002; 106: 1777–1782.
Trompet S, de Craen AJ, Postmus I et al: Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med Genet 2011; 12: 131.
Shepherd J, Blauw GJ, Murphy MB et al: Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet 2002; 360: 1623–1630.
Pare G, Chasman DI, Kellogg M et al: Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet 2008; 4: e1000118.
Clayton D : Testing for association on the X chromosome. Biostatistics 2008; 9: 593–600.
Marchini J, Howie B, Myers S, McVean G, Donnelly P : A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913.
Servin B, Stephens M : Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 2007; 3: e114.
Li Y, Ding J, Abecasis GR : Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006; 79: S2290.
Devlin B, Roeder K, Wasserman L : Genomic control, a new approach to genetic-based association studies. Theor Popul Biol 2001; 60: 155–166.
Gallinaro L, Cattini MG, Sztukowska M et al: A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood 2008; 111: 3540–3545.
Van Schie MC, Wieberdink RG, Koudstaal PJ et al: Genetic determinants of von Willebrand factor plasma levels and the risk of stroke: the Rotterdam Study. J Thromb Haemost 2012; 10: 550–556.
Desch KC, Ozel AB, Siemieniak D et al: Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc Natl Acad Sci USA 2013; 110: 588–593.
van Schie MC, de Maat MP, Isaacs A et al: Variation in the von Willebrand factor gene is associated with von Willebrand factor levels and with the risk for cardiovascular disease. Blood 2011; 117: 1393–1399.
van Schie MC, van Loon JE, de Maat MP, Leebeek FW : Genetic determinants of von Willebrand factor levels and activity in relation to the risk of cardiovascular disease: a review. J Thromb Haemost 2011; 9: 899–908.
Campos M, Sun W, Yu F et al: Genetic determinants of plasma von Willebrand factor antigen levels: a target gene SNP and haplotype analysis of ARIC cohort. Blood 2011; 117: 5224–5230.
Lowenstein CJ, Morrell CN, Yamakuchi M : Regulation of Weibel–Palade body exocytosis. Trends Cardiovasc Med 2005; 15: 302–308.
van Loon JE, Leebeek FW, Deckers JW et al: Effect of genetic variations in syntaxin-binding protein-5 and syntaxin-2 on von Willebrand factor concentration and cardiovascular risk. Circ Cardiovasc Genet 2010; 3: 507–512.
Harris EN, Weigel PH : The ligand-binding profile of HARE: hyaluronan and chondroitin sulfates A, C, and D bind to overlapping sites distinct from the sites for heparin, acetylated low-density lipoprotein, dermatan sulfate, and CS-E. Glycobiology 2008; 18: 638–648.
Komatsu M, Chiba T, Tatsumi K et al: A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 2004; 23: 1977–1986.
Daniel J, Liebau E : The ufm1 cascade. Cells 2014; 3: 627–638.
Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE : Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol 2006; 291: H1411–H1420.
Smith NL, Chen MH, Dehghan A et al: Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation 2010; 121: 1382–1392.
van Loon JE, Sanders YV, de Wee EM, Kruip MJ, de Maat MP, Leebeek FW : Effect of genetic variation in STXBP5 and STX2 on von Willebrand factor and bleeding phenotype in type 1 von Willebrand disease patients. PLoS One 2012; 7: e40624.
Acknowledgements
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute Contract U01HG004402; and National Institutes of Health Contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
We acknowledge the use of phenotype and genotype data from the B58C DNA collection, funded by Medical Research Council Grant G0000934 and the Wellcome Trust Grant 068545/Z/02 (http://www.b58cgene.sgul.ac.uk/).
Genotyping of the Lothian Birth Cohorts 1921 and 1936 were supported by the UK’s Biotechnology and Biological Sciences Research Council (BBSRC). Phenotype collection in the Lothian Birth Cohort 1921 was supported by The Chief Scientist Office of the Scottish Government (ETM/55). Phenotype collection in the Lothian Birth Cohort 1936 was supported by Research Into Ageing (continues as part of Age UK’s The Disconnected Mind project). The work was undertaken in The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Funding from the UK’s BBSRC, EPSRC, ESRC and MRC is gratefully acknowledged.
PROSPER is supported by the Scottish Executive Chief Scientist Office, Health Services Research Committee grant number CZG/4/306 and the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NOW 911-03-016).
The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Netherlands Heart Foundation (DHF-2007B159); the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO; 175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was further supported by the Netherlands Genomics Initiative (NGI)/NWO Project No. 050-060-810 and NWO/ZonMw Grant No. 918-76-619 and NWO/ZonMw Grant No. 918-76-619. Dr Dehghan is supported by NWO Grant (veni, 916.12.154) and the EUR Fellowship.
PREVEND genetics is supported by the Dutch Kidney Foundation (Grant E033), the EU project grant GENECURE (FP-6 LSHM CT 2006 037697), the National Institutes of Health (Grant 2R01LM010098), The Netherlands Organization for Health Research and Development (NWO-Groot Grant 175.010.2007.006, NWO VENI Grant 916.761.70, ZonMw Grant 90.700.441) and the Dutch Inter-University Cardiology Institute Netherlands (ICIN).
The VIS study in the Croatian island of Vis was supported through grants from the Medical Research Council UK and Ministry of Science, Education and Sport of the Republic of Croatia (No. 108-1080315-0302) and the European Union framework program 6 EUROSPAN project (Contract No. LSHG-CT-2006-018947). ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society and the European Union framework program 6 EUROSPAN project (Contract No. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Psaty serves on a DSMB for a clinical trial of a device funded by Zoll LifeCor. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
van Loon, J., Dehghan, A., Weihong, T. et al. Genome-wide association studies identify genetic loci for low von Willebrand factor levels. Eur J Hum Genet 24, 1035–1040 (2016). https://doi.org/10.1038/ejhg.2015.222
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2015.222
This article is cited by
-
Associations between blood type and COVID-19 infection, intubation, and death
Nature Communications (2020)
-
Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR
Nature Communications (2019)
-
Current and Emerging Options for the Management of Inherited von Willebrand Disease
Drugs (2017)