Abstract
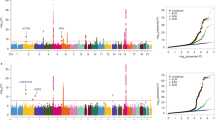
Recent genome-wide association studies have identified common variants at multiple loci influencing lung cancer risk. To decipher the genetic basis of the association signals at 3q28, 5p15.33, 6p21.33, 9p21 and 12p13.33, we performed a meta-analysis of data from five genome-wide association studies in populations of European ancestry totalling 12 316 lung cancer cases and 16 831 controls using imputation to recover untyped genotypes. For four of the regions, it was possible to refine the association signal identifying a smaller region of interest likely to harbour the functional variant. Our analysis did not provide evidence that any of the associations at the loci being a consequence of synthetic associations rather than linkage disequilibrium with a common risk variant at these risk loci.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM : Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Barker K, Martinez A, Wang R et al: PTEN mutations are uncommon in Proteus syndrome. J Med Genet 2001; 38: 480–481.
Hung RJ, McKay JD, Gaborieau V et al: A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008; 452: 633–637.
Amos CI, Wu X, Broderick P et al: Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008; 40: 616–622.
Thorgeirsson TE, Geller F, Sulem P et al: A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452: 638–642.
McKay JD, Hung RJ, Gaborieau V et al: Lung cancer susceptibility locus at 5p15.33. Nat Genet 2008; 40: 1404–1406.
Wang Y, Broderick P, Webb E et al: Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 2008; 40: 1407–1409.
Hu Z, Wu C, Shi Y et al: A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011; 43: 792–796.
Miki D, Kubo M, Takahashi A et al: Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet 2010; 42: 893–896.
Lan Q, Hsiung CA, Matsuo K et al: Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 2012; 44: 1330–1335.
Wang Y, Broderick P, Matakidou A, Vijayakrishnan J, Eisen T, Houlston RS : Variation in TP63 is associated with lung adenocarcinoma in the UK population. Cancer Epidemiol Biomark Prev 2011; 20: 1453–1462.
Wang Y, McKay JD, Rafnar T et al: Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 2014; 46: 736–741.
Travis WD, Brambilla E, Noguchi M et al: International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc 2011; 8: 381–385.
Broderick P, Wang Y, Vijayakrishnan J et al: Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res 2009; 69: 6633–6641.
Landi MT, Chatterjee N, Yu K et al: A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009; 85: 679–691.
Timofeeva MN, Hung RJ, Rafnar T et al: Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet 2012; 21: 4980–4995.
Shi J, Chatterjee N, Rotunno M et al: Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov 2012; 2: 131–139.
Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB : Rare variants create synthetic genome-wide associations. PLoS Biol 2010; 8: e1000294.
Su L, Zhou W, Asomaning K et al: Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis 2006; 27: 1024–1029.
Howie BN, Donnelly P, Marchini J : A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR : MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR : Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–959.
Zeggini E, Scott LJ, Saxena R et al: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638–645.
Marchini J, Howie B : Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511.
Aulchenko YS, Struchalin MV, van Duijn CM : ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinform 2010; 11: 134.
Dilthey AT, Moutsianas L, Leslie S, McVean G : HLA*IMP – an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 2011; 27: 968–972.
Leslie S, Donnelly P, McVean G : A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet 2008; 82: 48–56.
Clayton DG, Walker NM, Smyth DJ et al: Population structure, differential bias and genomic control in a large-scale, case–control association study. Nat Genet 2005; 37: 1243–1246.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Higgins JP, Thompson SG, Deeks JJ, Altman DG : Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560.
Bhattacharjee S, Rajaraman P, Jacobs KB et al: A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet 2012; 90: 821–835.
Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI : SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939.
Gabriel SB, Schaffner SF, Nguyen H et al: The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
Ward LD, Kellis M : HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids Res 2012; 40: D930–D934.
Boyle AP, Hong EL, Hariharan M et al: Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797.
Cooper GM, Stone EA, Asimenos G et al: Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005; 15: 901–913.
Shi J, Marconett CN, Duan J et al: Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun 2014; 5: 3365.
Willer CJ, Li Y, Abecasis GR : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
Lawrence MS, Stojanov P, Polak P et al: Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218.
Fletcher O, Houlston RS : Architecture of inherited susceptibility to common cancer. Nat Rev Cancer 2010; 10: 353–361.
Kote-Jarai Z, Saunders EJ, Leongamornlert DA et al: Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet 2013; 22: 2520–2528.
Bojesen SE, Pooley KA, Johnatty SE et al: Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 2013; 45: 371–384.
Walsh KM, Codd V, Smirnov IV et al: Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet 2014; 46: 731–735.
Bailey-Wilson JE, Amos CI, Pinney SM et al: A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am J Hum Genet 2004; 75: 460–474.
Acknowledgements
This study was supported by grants from the National Institute of Health (NIH) (U19CA148127, 5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197, R01 CA111703 and UO1 CA63673, R01 CA092039, CA074386, CA092824, CA090578), Cancer Research UK (C1298/A8780, C1298/A8362), HEAL, Sanofi-Aventis, Institut National du Cancer, the European Community (Integrated Project DNA repair, LSHG-CT-2005-512113), the Norwegian Cancer Association, the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101), the Fred Hutchinson Cancer Research Center, an FP7 grant (REGPOT 245536), the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure's Roadmap. We thank all individuals who participated in this study. The ICR study made use of data from the WTCCCII (http://www.wtccc.org.uk). We acknowledge the TCGA for their contribution of lung cancer genomic data to this study (Project Number 3230).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Wang, Y., Wei, Y., Gaborieau, V. et al. Deciphering associations for lung cancer risk through imputation and analysis of 12 316 cases and 16 831 controls. Eur J Hum Genet 23, 1723–1728 (2015). https://doi.org/10.1038/ejhg.2015.48
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2015.48