Abstract
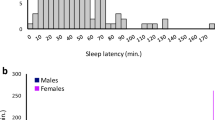
Time to fall asleep (sleep latency) is a major determinant of sleep quality. Chronic, long sleep latency is a major characteristic of sleep-onset insomnia and/or delayed sleep phase syndrome. In this study we aimed to discover common polymorphisms that contribute to the genetics of sleep latency. We performed a meta-analysis of genome-wide association studies (GWAS) including 2 572 737 single nucleotide polymorphisms (SNPs) established in seven European cohorts including 4242 individuals. We found a cluster of three highly correlated variants (rs9900428, rs9907432 and rs7211029) in the RNA-binding protein fox-1 homolog 3 gene (RBFOX3) associated with sleep latency (P-values=5.77 × 10−08, 6.59 × 10−08 and 9.17 × 10−08). These SNPs were replicated in up to 12 independent populations including 30 377 individuals (P-values=1.5 × 10−02, 7.0 × 10−03 and 2.5 × 10−03; combined meta-analysis P-values=5.5 × 10−07, 5.4 × 10−07 and 1.0 × 10−07). A functional prediction of RBFOX3 based on co-expression with other genes shows that this gene is predominantly expressed in brain (P-value=1.4 × 10−316) and the central nervous system (P-value=7.5 × 10−321). The predicted function of RBFOX3 based on co-expression analysis with other genes shows that this gene is significantly involved in the release cycle of neurotransmitters including gamma-aminobutyric acid and various monoamines (P-values<2.9 × 10−11) that are crucial in triggering the onset of sleep. To conclude, in this first large-scale GWAS of sleep latency we report a novel association of variants in RBFOX3 gene. Further, a functional prediction of RBFOX3 supports the involvement of RBFOX3 with sleep latency.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Krauchi K, Wirz-Justice A : Circadian clues to sleep onset mechanisms. Neuropsychopharmacology 2001; 25: S92–S96.
Cirelli C : The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci 2009; 10: 549–560.
Hastings M : The brain, circadian rhythms, and clock genes. BMJ 1998; 317: 1704–1707.
Hastings MH : Central clocking. Trends Neurosci 1997; 20: 459–464.
Baekeland F, Hoy P : Reported vs recorded sleep characteristics. Arch Gen Psychiatry 1971; 24: 548–551.
Hoch CC, Reynolds CF 3rd, Kupfer DJ, Berman SR, Houck PR, Stack JA : Empirical note: self-report versus recorded sleep in healthy seniors. Psychophysiology 1987; 24: 293–299.
Lehnkering H, Siegmund R : Influence of chronotype, season, and sex of subject on sleep behavior of young adults. Chronobiol Int 2007; 24: 875–888.
Geisler P, Tracik F, Cronlein T et al: The influence of age and sex on sleep latency in the MSLT-30—a normative study. Sleep 2006; 29: 687–692.
Krishnan V, Collop NA : Gender differences in sleep disorders. Curr Opin Pulm Med 2006; 12: 383–389.
Bonnet MH, Webb WB : The return to sleep. Biol Psychol 1979; 8: 225–233.
Afaghi A, O'Connor H, Chow CM : High-glycemic-index carbohydrate meals shorten sleep onset. Am J Clin Nutr 2007; 85: 426–430.
Nixon GM, Thompson JM, Han DY et al: Falling asleep: the determinants of sleep latency. Arch Dis Child 2009; 94: 686–689.
Gupta R, Dahiya S, Bhatia MS : Effect of depression on sleep: qualitative or quantitative? Indian J Psychiatry 2009; 51: 117–121.
Blader JC, Koplewicz HS, Abikoff H, Foley C : Sleep problems of elementary school children. A community survey. Arch Pediatr Adolesc Med 1997; 151: 473–480.
Eddy M, Walbroehl GS : Insomnia. Am Fam Physician 1999; 59: 1911–1916–1918.
Weitzman ED, Czeisler CA, Coleman RM et al: Delayed sleep phase syndrome. A chronobiological disorder with sleep-onset insomnia. Arch Gen Psychiatry 1981; 38: 737–746.
Freedman RR : EEG power spectra in sleep-onset insomnia. Electroencephalogr Clin Neurophysiol 1986; 63: 408–413.
van den Heuvel CJ, Lushington K : Chronobiology and insomnia: pathophysiology and treatment of circadian rhythm sleep disorders. Expert Rev Neurother 2002; 2: 249–260.
Touchette E, Petit D, Seguin JR, Boivin M, Tremblay RE, Montplaisir JY : Associations between sleep duration patterns and behavioral/cognitive functioning at school entry. Sleep 2007; 30: 1213–1219.
Heath AC, Kendler KS, Eaves LJ, Martin NG : Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep 1990; 13: 318–335.
Klei L, Reitz P, Miller M et al: Heritability of morningness-eveningness and self-report sleep measures in a family-based sample of 521 hutterites. Chronobiol Int 2005; 22: 1041–1054.
Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H : Genetic and environmental determination of human sleep. Sleep 1983; 6: 179–185.
Kantermann T, Juda M, Merrow M, Roenneberg T : The human circadian clock's seasonal adjustment is disrupted by daylight saving time. Curr Biol 2007; 17: 1996–2000.
DeMartinis NA, Winokur A : Effects of psychiatric medications on sleep and sleep disorders. CNS Neurol Disord Drug Targets 2007; 6: 17–29.
Wittmann M, Dinich J, Merrow M, Roenneberg T : Social jetlag: misalignment of biological and social time. Chronobiol Int 2006; 23: 497–509.
Li Y, Abecasis GR : Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006; S79: 2290.
Li Y, Willer C, Sanna S, Abecasis G : Genotype imputation. Annu Rev Genomics Hum Genet 2009; 10: 387–406.
Howie BN, Donnelly P, Marchini J : A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Marchini J, Howie B, Myers S, McVean G, Donnelly P : A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913.
Almasy L, Blangero J : Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211.
Liu F, Kirichenko A, Axenovich TI, van Duijn CM, Aulchenko YS : An approach for cutting large and complex pedigrees for linkage analysis. Eur J Hum Genet 2008; 16: 854–860.
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM : GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007; 23: 1294–1296.
Amin N, van Duijn CM, Aulchenko YS : A genomic background based method for association analysis in related individuals. PLoS One 2007; 2: e1274.
Aulchenko YS, Struchalin MV, van Duijn CM : ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 2010; 11: 134.
Wellcome Trust : Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–678.
Chen WM, Abecasis GR : Family-based association tests for genomewide association scans. Am J Hum Genet 2007; 81: 913–926.
Orwoll E, Blank JB, Barrett-Connor E et al: Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp Clin Trials 2005; 26: 569–585.
Zaykin DV : Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J Evol Biol 2011; 24: 1836–1841.
Fehrmann RS, Karjalainen JM, Krajewska M et al: Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet 2015; 47: 115–125.
Alter O, Brown PO, Botstein D : Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA 2000; 97: 10101–10106.
Sherlock G : Analysis of large-scale gene expression data. Brief Bioinform 2001; 2: 350–362.
Creyghton MP, Cheng AW, Welstead GG et al: Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 2010; 107: 21931–21936.
Kim KK, Adelstein RS, Kawamoto S : Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem 2009; 284: 31052–31061.
Dredge BK, Jensen KB : NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One 2011; 6: e21585.
Kiehl TR, Shibata H, Vo T, Huynh DP, Pulst SM : Identification and expression of a mouse ortholog of A2BP1. Mamm Genome 2001; 12: 595–601.
Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL : Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 2005; 25: 10005–10016.
McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA : A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol 2005; 5: 14.
Modrek B, Resch A, Grasso C, Lee C : Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res 2001; 29: 2850–2859.
Yeo G, Holste D, Kreiman G, Burge CB : Variation in alternative splicing across human tissues. Genome Biol 2004; 5: R74.
Morin LP, Hefton S, Studholme KM : Neurons identified by NeuN/Fox-3 immunoreactivity have a novel distribution in the hamster and mouse suprachiasmatic nucleus. Brain Res 2011; 1421: 44–51.
Bhalla K, Phillips HA, Crawford J et al: The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet 2004; 49: 308–311.
Barnby G, Abbott A, Sykes N et al: Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet 2005; 76: 950–966.
Martin CL, Duvall JA, Ilkin Y et al: Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 869–876.
Sebat J, Lakshmi B, Malhotra D et al: Strong association of de novo copy number mutations with autism. Science 2007; 316: 445–449.
Utami KH, Hillmer AM, Aksoy I et al: Detection of chromosomal breakpoints in patients with developmental delay and speech disorders. PLoS One 2014; 9: e90852.
Luppi PH : Neurochemical aspects of sleep regulation with specific focus on slow-wave sleep. World J Biol Psychiatry 2010; 11 (Suppl 1): 4–8.
Whiting PJ : The GABA-A receptor gene family: new targets for therapeutic intervention. Neurochem Int 1999; 34: 387–390.
Whiting PJ, Bonnert TP, McKernan RM et al: Molecular and functional diversity of the expanding GABA-A receptor gene family. Ann N Y Acad Sci 1999; 868: 645–653.
Buhr A, Bianchi MT, Baur R et al: Functional characterization of the new human GABA(A) receptor mutation beta3(R192H). Hum Genet 2002; 111: 154–160.
Charney DS, Nestler EJ : Neurobiology of mental illness. Oxford University Press: Oxford; New York, NY, USA, 2004.
Acknowledgements
We thank all the participants and staff of all the studies for their co-operation and contribution. We are grateful to all participants and their relatives, general practitioners and neurologists for their contributions and to P Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work and P Snijders for his help in data collection for the ERF study. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V Struchalin for their support in creation and analysis of imputed data for the Rotterdam Study. We are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. We thank Dr Fernando Rivadeneira, Dr Tobias A Knoch, Anis Abuseiris, Luc V de Zeeuw and Rob de Graaf for their help in creating GRIMP, and BigGRID, MediGRID and Services@MediGRID/D-Grid. We acknowledge the invaluable contributions of the recruitment team from the Croatian Centre for Global Health, University of Split, the administrative teams in Croatia and Edinburgh and the people of Split. Our work was supported by the FP6 programme EUCLOCK, the Dutch Science Foundation (the NWO), the Hersenstichting Nederland, the Rosalind Franklin Fellowships of the University of Groningen, targeted Financing from the Estonian Government, the European Union through the European Regional Development Fund in the frame of Centre of Excellence in Genomics, FP7 Projects ECOGENE, BBMRI, ENGAGE and OPENGENE, the Geestkracht programme of the Dutch Scientific Organization (ZON-MW) and matching funds from participating universities and mental health care organizations, the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, the EUROSPAN (European Special Populations Research Network) through the European Commission FP6 STRP grant, the Chief Scientist Office of the Scottish Government, the Royal Society, the Medical Research Council, Erasmus University, Erasmus MC, the Centre for Medical Systems Biology (CMSB1 and CMSB2) and the Netherlands Genomics Initiative (NGI) and ALBAN. We acknowledge the High Performance Computing Center of University of Tartu, for space and facilities to conduct part of the EGCUT analysis. We also acknowledge the Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), The Ministry of Education, Culture and Science, the Ministry of Health, Welfare and Sports of the Netherlands, the European Commission (DG XII) and the Municipality of Rotterdam. This study was supported by grants from the Netherlands Organization for Scientific Research (NWO-VENI grant 916.10.135 to LF) and a Horizon Breakthrough grant from the Netherlands Genomics Initiative (grant 92519031 to LF). The research leading to these results has received funding from the European Community’s Health Seventh Framework Programme (FP7/2007–2013) under grant agreement 259867. Study-specific acknowledgments and the ethical statement are provided in the Supplementary text.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Najaf Amin is supported by the Netherlands Brain Foundation (project number F2013(1)-28). Dr Gregory J Tranah was supported by NIA grant R01AG030474. Dr Henning Tiemeier was supported by the Vidi Grant of ZonMw (the Netherlands Organization for Health Research and Development, 2009-017.106.370). All other authors report no biomedical financial interests or potential conflicts of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Amin, N., Allebrandt, K., van der Spek, A. et al. Genetic variants in RBFOX3 are associated with sleep latency. Eur J Hum Genet 24, 1488–1495 (2016). https://doi.org/10.1038/ejhg.2016.31
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2016.31
This article is cited by
-
Genome-wide association in Drosophila identifies a role for Piezo and Proc-R in sleep latency
Scientific Reports (2024)
-
Genetics of circadian rhythms and sleep in human health and disease
Nature Reviews Genetics (2023)
-
The End of Snoring? Application of CRISPR/Cas9 Genome Editing for Sleep Disorders
Sleep and Vigilance (2018)
-
Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits
Nature Genetics (2017)