Abstract
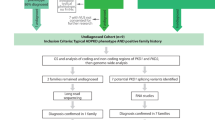
Autosomal dominant polycystic kidney disease (ADPKD) is the most common monogenic kidney disorder and is due to disease-causing variants in PKD1 or PKD2. Strong genotype–phenotype correlation exists although diagnostic sequencing is not part of routine clinical practice. This is because PKD1 bears 97.7% sequence similarity with six pseudogenes, requiring laborious and error-prone long-range PCR and Sanger sequencing to overcome. We hypothesised that whole-genome sequencing (WGS) would be able to overcome the problem of this sequence homology, because of 150 bp, paired-end reads and avoidance of capture bias that arises from targeted sequencing. We prospectively recruited a cohort of 28 unique pedigrees with ADPKD phenotype. Standard DNA extraction, library preparation and WGS were performed using Illumina HiSeq X and variants were classified following standard guidelines. Molecular diagnosis was made in 24 patients (86%), with 100% variant confirmation by current gold standard of long-range PCR and Sanger sequencing. We demonstrated unique alignment of sequencing reads over the pseudogene-homologous region. In addition to identifying function-affecting single-nucleotide variants and indels, we identified single- and multi-exon deletions affecting PKD1 and PKD2, which would have been challenging to identify using exome sequencing. We report the first use of WGS to diagnose ADPKD. This method overcomes pseudogene homology, provides uniform coverage, detects all variant types in a single test and is less labour-intensive than current techniques. This technique is translatable to a diagnostic setting, allows clinicians to make better-informed management decisions and has implications for other disease groups that are challenged by regions of confounding sequence homology.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cornec-Le Gall E, Audrezet MP, Chen JM et al: Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol 2013; 24: 1006–1013.
Cornec-Le Gall E, Audrézet M-P, Le Meur Y, Chen J-M, Férec C : Genetics and pathogenesis of autosomal dominant polycystic kidney disease: 20 years on. Hum Mutat 2014; 35: 1393–1406.
Kirsch S, Pasantes J, Wolf A et al: Chromosomal evolution of the PKD1 gene family in primates. BMC Evol Biol 2008; 8: 263.
Symmons O, Varadi A, Aranyi T : How segmental duplications shape our genome: recent evolution of ABCC6 and PKD1 Mendelian disease genes. Mol Biol Evol 2008; 25: 2601–2613.
Rossetti S, Hopp K, Sikkink RA et al: Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. J Am Soc Nephrol 2012; 23: 915–933.
Rossetti S, Strmecki L, Gamble V, Komel R, Winearls CG : Mutation analysis of the entire PKD1 gene: genetic and diagnostic implications. Am J Hum Genet 2001; 68: 46–63.
Tan AY, Blumenfeld J, Michaeel A et al: Autosomal dominant polycystic kidney disease caused by somatic and germline mosaicism. Clin Genet 2014; 87: 373–377.
Rossetti S, Consugar MB, Chapman AB et al: Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2007; 18: 2143–2160.
Audrézet M-P, Cornec-Le Gall E, Chen J-M et al: Autosomal dominant polycystic kidney disease: comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat 2012; 33: 1239–1250.
Tan AY, Michaeel A, Liu G et al: Molecular diagnosis of autosomal dominant polycystic kidney disease using next-generation sequencing. J Mol Diagn 2014; 16: 216–228.
Trujillano D, Bullich G, Ossowski S et al: Diagnosis of autosomal dominant polycystic kidney disease using efficient PKD1and PKD2targeted next-generation sequencing. Mol Genet Genomic Med 2014; 2: 412–421.
Eisenberger T, Decker C, Hiersche M et al: An efficient and comprehensive strategy for genetic diagnostics of polycystic kidney disease. PLoS One 2015; 10: e0116680.
Meynert A, Ansari M, FitzPatrick D, Taylor M : Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics 2014; 15: 247–258.
Biesecker LG, Green RC : Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014; 370: 2418–2425.
Nishiguchi K, Tearle R, Liu Y, Oh E, Katsanis N, Rivolta C : Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc Natl Acad Sci USA 2013; 10: 16139–16144.
Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347.
Pei Y, Obaji J, Dupuis A et al: Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol 2009; 20: 205–212.
Harris PC, Torres VE : Polycystic kidney disease, autosomal dominant; in Pagon RA, Adam MP, Ardinger HH (eds): Seattle: University of Washington, 2015. Available at http://www.ncbi.nlm.nih.gov/books/NBK1246/.
Li H . Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013. Available at http://arxiv.org/abs/1303.3997.
DePristo MA, Banks E, Poplin R et al: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498.
Illumina. Sequencing coverage calculation methods for human whole-genome sequencing. 2014:1–2. Available at http://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/hiseq-x-30x-coverage-technical-note-770-2014-042.pdf.
Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424.
ADPKD Mutation Database: PKDB. Available at http://pkdb.pkdcure.org (accessed 30 April 2015).
Layer RM, Chiang C, Quinlan AR, Hall IM : LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 2014; 15: 1–19.
Abyzov A, Urban AE, Snyder M, Gerstein M : CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 2011; 21: 974–984.
Chiang C, Layer RM, Faust GG et al: SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods 2015; 12: 966–968.
MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW : The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2013; 42: D986–D992.
Kingsmore S, Saunders C : Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Sci Transl Med 2011; 3: 23–27.
Aird D, Ross MG, Chen W-S et al: Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 2011; 12: R18.
Bogdanova N, Markoff A, Gerke V, McCluskey M, Horst J, Dworniczak B : Homologues to the first gene for autosomal dominant polycystic kidney disease are pseudogenes. Genomics 2001; 74: 333–341.
Simms R, Travis D, Durkie M, Wilson G, Dalton A, Ong A : Genetic testing in the assessment of living related kidney donors at risk of autosomal dominant polycystic kidney disease. Transplantation 2014; 99: 1023–1029.
Acknowledgements
Many thanks for the patients and families that participated in the study. MJC is supported by a Cancer Institute NSW early career fellowship (13/ECF/1-46). We acknowledge financial support from the Burcher, Kirkpatrick and Lewis families, and the Kinghorn Foundation, without which this research would not have been possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Mallawaarachchi, A., Hort, Y., Cowley, M. et al. Whole-genome sequencing overcomes pseudogene homology to diagnose autosomal dominant polycystic kidney disease. Eur J Hum Genet 24, 1584–1590 (2016). https://doi.org/10.1038/ejhg.2016.48
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2016.48
This article is cited by
-
Atypical splicing variants in PKD1 explain most undiagnosed typical familial ADPKD
npj Genomic Medicine (2023)
-
Identification of novel single-nucleotide variants altering RNA splicing of PKD1 and PKD2
Journal of Human Genetics (2022)
-
Recommendations for whole genome sequencing in diagnostics for rare diseases
European Journal of Human Genetics (2022)
-
Identification of ACOT13 and PTGER2 as novel candidate genes of autosomal dominant polycystic kidney disease through whole exome sequencing
European Journal of Medical Research (2021)
-
Genomic diagnostics in polycystic kidney disease: an assessment of real-world use of whole-genome sequencing
European Journal of Human Genetics (2021)