Abstract
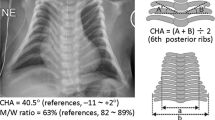
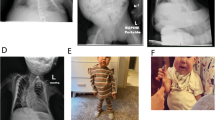
In approximately 20% of individuals with Kagami-Ogata syndrome (KOS14, MIM 608149), characterized by a bell-shaped thorax with coat-hanger configuration of the ribs, joint contractures, abdominal wall defects and polyhydramnios during the pregnancy, the syndrome is caused by a maternal deletion of the imprinted gene cluster in chromosome 14q32.2. Most deletions reported so far included one or both of the differentially methylated regions (DMRs) – DLK1/MEG3 IG-DMR and MEG3-DMR. We present two unrelated families with two affected siblings each, presenting with classical KOS14 due to maternally inherited microdeletions. Interestingly, all four patients have lived through to adulthood, even though mortality rates for patients with KOS14 due to a microdeletion are relatively high. In the first family, none of the DMRs is included in the deletion and the methylation status is identical to that of controls. Deletions that do not encompass the DMRs in this region are thus sufficient to elicit the full KOS14 phenotype. In the second family, a partially overlapping deletion including both DMRs and MEG3 was detected. In summary, we show that patients with KOS14 can live into adulthood, that causal deletions do not have to include the DMRs and that consequently a normal methylation pattern does not exclude KOS14.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lin SP, Youngson N, Takada S et al: Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 2003; 35: 97–102.
Paulsen M, Takada S, Youngson NA et al: Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res 2001; 11: 2085–2094.
Kagami M, Sekita Y, Nishimura G et al: Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008; 40: 237–242.
Kagami M, O'Sullivan MJ, Green AJ et al: The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 2010; 6: e1000992.
Beygo J, Elbracht M, de Groot K et al: Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32. Eur J Hum Genet 2015; 23: 180–188.
Sutton VR, Shaffer LG : Search for imprinted regions on chromosome 14: comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am J Med Genet 2000; 93: 381–387.
Kagami M, Nishimura G, Okuyama T et al: Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am J Med Genet A 2005; 138A: 127–132.
Kagami M, Kato F, Matsubara K, Sato T, Nishimura G, Ogata T : Relative frequency of underlying genetic causes for the development of UPD(14)pat-like phenotype. Eur J Hum Genet 2012; 20: 928–932.
Ogata T, Kagami M : Kagami-Ogata syndrome: a clinically recognizable upd(14)pat and related disorder affecting the chromosome 14q32.2 imprinted region. J Hum Genet 2015; 61: 87–94.
Vandeweyer G, Reyniers E, Wuyts W, Rooms L, Kooy RF : CNV-WebStore: online CNV analysis, storage and interpretation. BMC Bioinformatics 2011; 12: 4.
Beygo J, Citro V, Sparago A et al: The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum Molr Genet 2013; 22: 544–557.
D'Haene B, Vandesompele J, Hellemans J : Accurate and objective copy number profiling using real-time quantitative PCR. Methods 2010; 50: 262–270.
de Vree PJ, de Wit E, Yilmaz M et al: Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol 2014; 32: 1019–1025.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA : Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 2007; 35: W71–W74.
Kagami M, Kurosawa K, Miyazaki O, Ishino F, Matsuoka K, Ogata T : Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome). Eur J Hum Genet 2015; 23: 1488–1498.
Corsello G, Salzano E, Vecchio D et al: Paternal uniparental disomy chromosome 14-like syndrome due a maternal de novo 160kb deletion at the 14q32.2 region not encompassing the IG- and the MEG3-DMRs: Patient report and genotype-phenotype correlation. Am J Med Genet A 2015; 167A: 3130–3138.
Rosenfeld JA, Fox JE, Descartes M et al: Clinical features associated with copy number variations of the 14q32 imprinted gene cluster. Am J Med Genet A 2015; 167A: 345–353.
Lee JA, Carvalho CM, Lupski JR : A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 2007; 131: 1235–1247.
Liu P, Carvalho CM, Hastings P, Lupski JR : Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev 2012; 22: 211–220.
Kagami M, Matsuoka K, Nagai T et al: Paternal uniparental disomy 14 and related disorders: placental gene expression analyses and histological examinations. Epigenetics 2012; 7: 1142–1150.
Benetatos L, Hatzimichael E, Londin E et al: The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci 2013; 70: 795–814.
Acknowledgements
We thank Dr Max van Min for his advice on TLA sequencing, Christina Lich for technical assistance and Jasmin Beygo for her help with figures and helpful discussions. IMvdW is funded by the Special Research Fund of the University of Antwerp (Bijzonder Onderzoeksfonds (BOF-IWT)). GV is a postdoctoral fellow of the Research Fund Flanders (FWO). Part of the work was funded by the Bundesministerium für Bildung und Forschung (Network Imprinting diseases, 01GM1513A).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
van der Werf, I., Buiting, K., Czeschik, C. et al. Novel microdeletions on chromosome 14q32.2 suggest a potential role for non-coding RNAs in Kagami-Ogata syndrome. Eur J Hum Genet 24, 1724–1729 (2016). https://doi.org/10.1038/ejhg.2016.82
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2016.82
This article is cited by
-
Measurement of genetic diseases as a cause of mortality in infants receiving whole genome sequencing
npj Genomic Medicine (2020)
-
CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains
Genome Biology (2019)
-
Maternally inherited 133kb deletion of 14q32 causing Kagami–Ogata syndrome
Journal of Human Genetics (2018)
-
New insights into the imprinted MEG8-DMR in 14q32 and clinical and molecular description of novel patients with Temple syndrome
European Journal of Human Genetics (2017)