Abstract
Purpose
Autosomal-dominant optic atrophy (ADOA), often associated with mutations in the OPA1 gene (chromosome 3q28-q29) is rarely reported in Asia. Our aim was to identify and describe this condition in an Asian population in Singapore.
Patients and methods
Preliminary cross-sectional study at the Singapore National Eye Centre, including patients with clinical suspicion of ADOA, who subsequently underwent genetic testing by direct sequencing of the OPA1 gene.
Results
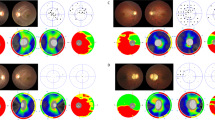
Among 12 patients (10 families) with clinically suspected ADOA, 7 patients (5 families) from 3 different ethnic origins (Chinese, Indian, and Malay) carried a heterozygous pathogenic variant in the OPA1 gene. The OPA1 mutations were located on exons 8, 9, 11, and 17: c.869G>A (p.Arg290Glu), c.892A>G (p.Ser298Gly), c.1140G>A (splicing mutation), and c.1669C>T (p.Arg557*), respectively. One splicing mutation (c.871-1G>A) was identified in intron 8. We also identified a novel mutation causing optic atrophy and deafness (c.892A>G (p.Ser298Gly)). Among the phenotypic features, colour pupillometry disclosed a dissociation between low vision and preserved pupillary light reflex in ADOA.
Conclusion
We report the first cases of genetically confirmed OPA1-related ADOA from Singapore, including a novel mutation causing ‘ADOA plus’ syndrome. Further epidemiological studies are needed in order to determine the prevalence of ADOA in South-East Asia.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lenaers G, Hamel C, Delettre CC, Amati-Bonneau P, Procaccio V, Bonneau D et al. Dominant optic atrophy. Orphanet J Rare Dis 2012; 7: 46–46.
Milea D, Amati-Bonneau P, Reynier P, Bonneau D . Genetically determined optic neuropathies. Curr Opin Neurol 2010; 23 (1): 24–28.
Delettre C, Lenaers G, Griffoin J-M, Gigarel N, Lorenzo C, Belenguer P et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 2000; 26 (2): 207–210.
Alexander C, Votruba M, Pesch UEA, Thiselton DL, Mayer S, Moore A et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet 2000; 26 (2): 211–215.
Chao de la Barca JM, Prunier-Mirebeau D, Amati-Bonneau P, Ferré M, Sarzi E, Bris C et al. OPA1-related disorders: diversity of clinical expression, modes of inheritance and pathophysiology. Neurobiol Dis 2016; 90: 20–26.
Ferré M, Amati-Bonneau P, Tourmen Y, Malthièry Y, Reynier P . eOPA1: an online database for OPA1 mutations. Hum Mutat 2005; 25 (5): 423–428.
Ferré M, Caignard A, Milea D, Leruez S, Cassereau J, Chevrollier A et al. Improved locus-specific database for OPA1 mutations allows inclusion of advanced clinical data. Hum Mutat 2015; 36 (1): 20–25.
Cohn AC, Toomes C, Potter C, Towns KV, Hewitt AW, Inglehearn CF et al. Autosomal dominant optic atrophy: penetrance and expressivity in patients with OPA1 mutations. Am J Ophthalmol 2007; 143 (4): 656–662.e651.
Ferré M, Bonneau D, Milea D, Chevrollier A, Verny C, Dollfus H et al. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 novel OPA1 mutations. Hum Mutat 2009; 30 (7): E692–E705.
Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissière A et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy 'plus' phenotypes. Brain 2008; 131 (2): 338–351.
Bonneau D, Colin E, Oca F, Ferré M, Chevrollier A, Guéguen N et al. Early-onset Behr syndrome due to compound heterozygous mutations in OPA1. Brain 2014; 137 (10): e301–e301.
Spiegel R, Saada A, Flannery PJ, Burté F, Soiferman D, Khayat M et al. Fatal infantile mitochondrial encephalomyopathy, hypertrophic cardiomyopathy and optic atrophy associated with a homozygous OPA1 mutation. J Med Genet 2016; 53 (2): 127–131.
Kjer P . Infantile optic atrophy with dominant mode of inheritance: a clinical and genetic study of 19 Danish families. Acta Ophthalmol Suppl 1959; 164 (Supp 54): 1–147.
Yu-Wai-Man P, Griffiths PG, Burke A, Sellar PW, Clarke MP, Gnanaraj L et al. The prevalence and natural history of dominant optic atrophy due to <em>OPA1</em> mutations. Ophthalmology 2010; 117 (8): 1538–1546.e1531.
Galvez-Ruiz A, Neuhaus C, Bergmann C, Bolz H . First cases of dominant optic atrophy in Saudi Arabia: report of two novel OPA1 mutations. J Neuroophthalmol 2013; 33 (4): 349–353.
Dadgar S, Hagens O, Dadgar SR, Haghighi EN, Schimpf S, Wissinger B et al. Structural model of the OPA1 GTPase domain may explain the molecular consequences of a novel mutation in a family with autosomal dominant optic atrophy. Exp Eye Res 2006; 83 (3): 702–706.
Liskova P, Ulmanova O, Tesina P, Melsova H, Diblik P, Hansikova H et al. Novel OPA1 missense mutation in a family with optic atrophy and severe widespread neurological disorder. Acta Ophthalmol 2013; 91 (3): e225–e231.
Hamahata T, Fujimaki T, Fujiki K, Miyazaki A, Mizota A, Murakami A . OPA1 mutations in Japanese patients suspected to have autosomal dominant optic atrophy. Jpn J Ophthalmol 2012; 56 (1): 91–97.
Yen M-Y, Wang A-G, Lin Y-C, Fann M-J, Hsiao K-J . Novel mutations of the OPA1 gene in Chinese dominant optic atrophy. Ophthalmology 2010; 117 (2): 392–396.e1.
Chen Y, Jia X, Wang P, Xiao X, Li S, Guo X et al. Mutation survey of the optic atrophy 1 gene in 193 Chinese families with suspected hereditary optic neuropathy. Mol Vis 2013; 19: 292–302.
Rukmini AV, Milea D, Baskaran M, How AC, Perera SA, Aung T et al. Pupillary responses to high-irradiance blue light correlate with glaucoma severity. Ophthalmology 2015; 122 (9): 1777–1785.
Toomes C, Marchbank NJ, Mackey DA, Craig JE, Newbury-Ecob RA, Bennett CP et al. Spectrum, frequency and penetrance of OPA1 mutations in dominant optic atrophy. Hum Mol Genet 2001; 10 (13): 1369–1378.
Leo-Kottler B, Luberichs J, Besch D, Christ-Adler M, Fauser S . Leber's hereditary optic neuropathy: clinical and molecular genetic results in a patient with a point mutation at np T11253C (isoleucine to threonine) in the ND4 gene and spontaneous recovery. Graefes Arch Clin Exp Ophthalmol 2002; 240 (9): 758–764.
Schimpf S, Schaich S, Wissinger B . Activation of cryptic splice sites is a frequent splicing defect mechanism caused by mutations in exon and intron sequences of the OPA1 gene. Hum Genet 2006; 118 (6): 767–771.
Zhang A-M, Bi R, Hu Q-X, Fan Y, Zhang Q, Yao Y-G . The OPA1 gene mutations are frequent in Han Chinese patients with suspected optic neuropathy. Mol Neurobiol 2016; 1–9.
Nissen C, Ronnback C, Sander B, Herbst K, Milea D, Larsen M et al. Dissociation of pupillary post-illumination responses from visual function in confirmed OPA1 c.983A >G and c.2708_2711delTTAG autosomal dominant optic atrophy. Front Neurol 2015; 6: 5.
Kawasaki A, Herbst K, Sander B, Milea D . Selective wavelength pupillometry in Leber hereditary optic neuropathy. Clin Exp Ophthalmol 2010; 38 (3): 322–324.
Leruez S, Milea D, Defoort-Dhellemmes S, Colin E, Crochet M, Procaccio V et al. Sensorineural hearing loss in OPA1-linked disorders. Brain 2013; 136 (7): e236–e236.
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A . Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010; 20 (1): 110–121.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. Fast, scalable generation of high‚ quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539.
Schwarz JM, Cooper DN, Schuelke M, Seelow D . MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11 (4): 361–362.
Ng PC, Henikoff S . Predicting deleterious amino acid substitutions. Genome Res 2001; 11 (5): 863–874.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7 (4): 248–249.
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29 (1): 308–311.
Exome Aggregation Consortium (ExAC), Cambridge, MA, USA. Available at: http://exac.broadinstitute.org (accessed on February 2016).
Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA. Available at: http://evs.gs.washington.edu/EVS/ (accessed on February 2016).
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012; 491 (7422): 56–65.
Acknowledgements
This work was supported by the National Medical Research Council, Singapore (Grant Nos: NMRC/CIRG/1401/2014(DM) and NMRC/NIG/1000/2009 (JJG)) and the Singapore National Eye Centre Health Research Endowment Fund, Singapore (Grant No: 1005/20/2013 (DM)). The funding organisations had no role in the design and conduct of the research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Loo, J., Singhal, S., Rukmini, A. et al. Multiethnic involvement in autosomal-dominant optic atrophy in Singapore. Eye 31, 475–480 (2017). https://doi.org/10.1038/eye.2016.255
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/eye.2016.255
This article is cited by
-
Basics, benefits, and pitfalls of pupillometers assessing visual function
Eye (2024)
-
Fast and deep phosphoproteome analysis with the Orbitrap Astral mass spectrometer
Nature Communications (2024)