Abstract
Purpose:
Treacher Collins syndrome is a mandibulofacial dysostosis caused by mutations in genes involved in ribosome biogenesis and synthesis. TCOF1 mutations are observed in ~80% of the patients and are inherited in an autosomal dominant manner. Recently, two other genes have been reported in <2% of patients—POLR1D in patients with autosomal dominant inheritance, and POLR1C in patients with autosomal recessive inheritance.
Methods:
We performed direct sequencing of TCOF1, POLR1C, and POLR1D in two unrelated consanguineous families.
Results:
The four affected children shared the same homozygous mutation in POLR1D (c.163C>G, p.Leu55Val). This mutation is localized in a region encoding the dimerization domain of the RNA polymerase. It is supposed that this mutation impairs RNA polymerase, resulting in a lower amount of mature dimeric ribosomes. A functional analysis of the transcripts of TCOF1 by real-time quantitative reverse transcription–polymerase chain reaction was performed in the first family, demonstrating a 50% reduction in the index case, compatible with this hypothesis.
Conclusion:
This is the first report of POLR1D mutation being responsible for an autosomal recessive inherited Treacher Collins syndrome. These results reinforce the concept of genetic heterogeneity of Treacher Collins syndrome and underline the importance of combining clinical expertise and familial molecular analyses for appropriate genetic counseling.
Genet Med 16 9, 720–724.
Similar content being viewed by others
Main
First reported in 1846 by Thomson1 and later in 1889 by Berry,2 Treacher Collins syndrome (TCS), also named Franceschetti–Zwahlen–Klein syndrome (OMIM 154500), was well defined by Treacher Collins in 1900.3 TCS belongs to a clinically and genetically heterogeneous group of mandibulofacial dysostoses. It occurs with an estimated incidence of 1/50,000 live births.4 This craniofacial disorder is characterized by a combination of bilateral hypoplasia of the facial bones, downward slanting of the palpebral fissures, colobomas of the lower eyelids, sparse eyelashes, cleft palate, and malformations of the external and the middle ears responsible for bilateral conductive hearing loss. TCS results from a bilateral and symmetrical default of growth of the structures derived from the first and second branchial arches.4
To date, three genes have been implicated: TCOF1 mutations account for ~80% of TCS patients and are inherited in an autosomal dominant manner, whereas mutations in the POLR1D and POLR1C genes are observed in <2%, with autosomal dominant and autosomal recessive inheritance patterns, respectively.5 These genes encode proteins that play a key role in ribosome biogenesis and synthesis. Consequently, TCS is considered to be a ribosomopathy.
We report on a new POLR1D autosomal recessive mutation in two unrelated Turkish consanguineous families.
Materials and Methods
Clinical reports
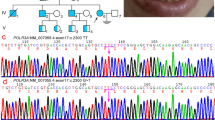
Family 1. The affected individual was a young girl, born from distant cousins who originated from the region of Cappadocia in Turkey. She was born at term after an uneventful pregnancy with normal growth parameters (weight: 3,700 g; height: 52 cm; head circumference: 33 cm). Microretrognathia, downslanting palpebral fissures, malar hypoplasia, and right-ear dysplasia with bilateral preauricular fistulae were observed ( Figure 1a ), but no diagnosis was proposed. When evaluated at the age of 3 years by a geneticist (B.D.), the diagnosis of TCS was suggested considering these clinical signs and the associated bilateral hearing loss. Magnetic resonance imaging and computed tomography scan of the skull showed bilateral hypoplasia of oval window, prolapsus of the second portion of the facial canal, and rudimentary auditory ossicles. Growth parameters were normal (at + 1SD), as were motor and cognitive development parameters. Clinical examination and radiological investigations (including skull X-rays and pantograms) of her parents and her sister were normal ( Figure 1b ). A maternal cousin, also born from a consanguineous union, had unilateral anotia, microretrognathism, and asymmetric mandibular hypoplasia consistent with a possible diagnosis of Goldenhar syndrome.
Photographs of Family 1 visualizing the affected individual and her unaffected parents and sister. (a) Frontal view of the patient at the ages of (1) 1 year, (2) 3 years, and (3) 7 years, showing typical features of Treacher Collins syndrome with microretrognathia, downslanting palpebral fissures, malar hypoplasia, right-ear dysplasia, and bilateral hearing loss. (b) Frontal view of her unaffected (1) father, (2) mother, and (3) sister. Written consent was obtained to publish photographs of all individuals shown herein.
Family 2. The three affected children were born from first cousins originating from the same region of central Turkey as Family 1. The elder sister was born at term after an unremarkable pregnancy. Her development was normal during the first years of life. She came to medical attention due to progressive learning difficulties and hearing loss at the age of 8 years. A 45- to 50-dB conductive deafness was diagnosed, covering all frequencies, and magnetic resonance imaging visualized a bilateral fusion between the uncus and the hammer. Her hearing recovered completely after microsurgery. These conditions, associated with malar hypoplasia and retrognathism, led the clinical geneticist (L.V.M.) to diagnose TCS.
At the time of diagnosis, her 5-year-old sister was examined, and her phenotype was considered normal. Five years later, a younger brother was born with severe micrognathia, cleft palate, downslanting palpebral fissures, and marked malar hypoplasia. Conductive deafness due to major ossicular hypoplasia was severe, requiring hearing aids. His course indicated behavioral problems, and special education was needed. When molecular investigation of recessive TCS became available, the family was reevaluated. Surprisingly, craniofacial dysmorphism and hearing loss were also observed in the allegedly normal sister, compatible with a TCS phenotype.
Molecular investigations
DNA extraction. DNA samples were obtained after receiving informed consent. Genomic DNA from blood was extracted using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany).
Sequencing analysis. Direct sequencing of the coding regions of the TCOF1, POLR1D, and POLR1C genes, as well as their intron–exon boundaries, was performed. In detail, polymerase chain reaction (PCR) products were purified using QIAamp (Qiagen) and were sequenced using a BigDye Terminator, version 3.1, Cycle Sequencing Kit (Life Technologies, Carlsbad, CA) according to the manufacturers’ instructions. The sequencing products were purified with Sephadex G50 in a 96-well format. Analyses of the forward and reverse sequences were performed on an ABI Prism 3130 Genetic Analyzer using Seqscape 2.6 software (Life Technologies), and these sequences were compared with the genomic reference sequence (TCOF1: NG_011341.1; POLR1D: NG_028908.1; POLR1C: NG_028283.1) for intronic regions and exon numbering. The reference of the complementary DNA (cDNA) corresponds to the major Treacle isoform of TCOF1 (NM_001135243.1).
Multiplex ligation-dependent probe amplification. Multiplex ligation-dependent probe amplification (MLPA) analysis was performed using the Salsa MLPA Kit P310-B2 TCOF1 (MRC-Holland, Amsterdam, The Netherlands). The procedure followed for MLPA analysis was that described by the manufacturer. Additional information about commercially available probe sets is available at http://www.mrc-holland.com. The products were then separated by capillary electrophoresis on an ABI 3130 DNA Analyzer (Life Technologies). The results were analyzed by GeneMapper software, version 4.0 (Life Technologies) and MRC Coffalyzer MLPA-Dat Software (MRC-Holland). A reduction or increase of 40% or more in peak area, as compared with normal controls, was considered suggestive of an aberrant amplification.
Quantitative real-time PCR of TCOF1 expression (Family 1 only). Total RNA was extracted from samples stored at −80 °C in PaxGene RNA collection tubes using a PaxGene Blood RNA Isolation Kit (PreAnalytiX, Qiagen/BD Company (Hombrechtikon, Switzerland)) according to the manufacturer’s recommendations. All reagents were obtained from Applied Biosystems (Carlsbad, CA). Briefly, 500 ng of total RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit according to the manufacturer’s instructions (Life Technologies). Quantitative real-time PCR was performed in triplicate using the StepOnePlus system (Applied Biosystems) and a TaqMan Universal PCR Master Mix with 1 μl cDNA input per plate (for a final volume of 20 µl) and 1.5 µl of Hs00494133_ml Taqman Gene Expression assay (corresponding to exons 2–3 of TCOF1) or Hs00947122_m1 TaqMan Gene Expression assay (corresponding to exons 17–18 of TCOF1) with 18S or hypoxanthine phosphoribosyltransferase TaqMan VIC Endogenous Controls. Cycle threshold (Ct) values were calculated using the StepOnePlus software, version 2, using automatic baseline settings and a threshold of 0.2. The Ct value of an endogenous control gene (18S or hypoxanthine phosphoribosyltransferase) was subtracted from the corresponding Ct value for the target gene, resulting in the ΔCt value that was used for relative quantification of RNA expression. The expression data of TCOF1 transcripts were determined by relative quantification in comparison with a pool of four RNA controls.
Haplotype analysis at the POLR1D locus. Haplotype analyses were performed on different members of the two families using polymorphic microsatellite DNA markers flanking POLR1D (position chr13:28194880–28241559): D13S1292, 135 kb; 43GT, 37 kb (chr13:28157007–28157693); D13S1244, 5.2 kb; and 19xGT, 48 kb (chr13:28289919–28290656).
Primer sequences for D2S marker amplifications were selected from the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) Database (http://genome.ucsc.edu); the others were designed by the primer-BLAST software of the National Center for Biotechnology Information. The amplifications were performed with TaqMan PCR master mix (Qiagen) using the characteristic melting temperature of primers. Alleles were separated on an ABI 3130 Genetic Analyzer (Life Technologies), and allele lengths were determined by GeneMapper software, version 4.0 (Life Technologies).
Results
TCOF1 sequencing results of the index patients from Families 1 and 2 were normal. No large rearrangement within the TCOF1 gene was found using MLPA. No POLR1C mutation was detected. POLR1D sequencing identified a homozygous c.163C>G mutation in the affected individuals of the two families, predicted to determine a p.Leu55Val residue change. Familial segregation study of this mutation demonstrated that the parents in both the families and the unaffected sister in Family 1 (II.1) were heterozygous. No DNA of the maternal cousin suspected of having Goldenhar syndrome was available for genetic studies. This mutation was not found in a series of 150 control chromosomes.
A 50% decrease in TCOF1 transcripts was observed by reverse transcription–PCR study in the leukocytes of the affected patient of Family 1. TCOF1 mRNA expression was normal in her parents and her unaffected sister ( Figure 2a ).
Localization and effect of the C.163C>G mutation. (a) Transcript levels of TCOF1 using quantitative real-time reverse transcription–polymerase chain reaction in leukocyte samples from individuals in Family 1 demonstrating 50% decrease of TCOF1 mRNA in the affected patient and normal levels of transcripts in her unaffected parents and sister. The error bars indicate ± SD. (b) Evolutionary conservation of the amino acid residues affected by missense mutations in the region within exon 3 of POLR1D (black arrows: mutations reported by Dauwerse et al.5; red arrows: mutation reported in the families studied herein).
Haplotype analysis revealed that both families had different microsatellite markers surrounding the POLR1D gene (Supplementary Figure S1 online).
Discussion
TCS has been considered an autosomal dominant condition for decades,6 its common sporadic occurrence being explained by a high rate of de novo TCOF1 mutations.7,8 However, recent identification of disease-causing mutations in POLR1C and POLR1D with autosomal recessive and autosomal dominant modes of inheritance pointed to heterogeneity in both the locus and the mode of inheritance.3
Treacle is a nuclear phosphoprotein. During embryogenesis, Treacle is expressed in the neuroepithelium that gives rise to the neural crest cells, the latter differentiating into cartilage, bones, and connective tissue of the head and the face.9 TCOF1 mutations reported so far exert their effect through a haploinsufficiency mechanism.10 Treacle loss of function alters ribosome biosynthesis in mature cells of the neuroepithelium and neural crest cells, thus affecting both formation and proliferation, thereby leading to abnormal development of the first and the second branchial arches.11 POLR1C and POLR1D genes encode RNA polymerases I and III (PolI/PolIII) subunits, which are the major players in 70S ribosome biosynthesis. Mutations in POLR1D/C genes induce a decrease in functional PolI (which colocalizes with Treacle)12 and PolIII, leading to a decreased number of mature ribosomes in the neuroepithelium and neural crest cells at a critical time of embryogenesis.3,11
All the 17 mutations identified so far in the POLR1D gene are located in exon 3 (five “frameshift,” four nonsense, seven missense, and one splice-site mutations). The mode of inheritance of all these mutations is autosomal dominant, with high incomplete penetrance.3
Our TCS families are the first POLR1D-mutated examples corresponding to the autosomal recessive mode of inheritance. All four affected patients are homozygous for an exon 3 c.163C>G missense mutation, converting a leucine to a valine at position 55. This novel mutation is observed neither in the 1000 Genomes database (http://www.1000genomes.org/data) nor in the Exome Variant Server (http://evs.gs.washington.edu). Leucine at position 55 is highly conserved across species. Similarly, the whole sequence appears to be conserved. Interestingly, this mutation is also located in exon 3 of the POLR1D gene and is placed within a hot spot of eight mutations between amino acids 45 and 57 ( Figure 2b ). Our haplotype studies support the hypothesis that this mutation in the two families arose from independent events and not from a founder effect, even though both families originated from the same geographic region. This cluster of mutations is located in a region of the protein that contains the RNA polymerase dimerization domain.13 We hypothesize that this mutation is not responsible for major modifications of the protein in the heterozygous state but that it makes the dimerization of the protein impossible in the homozygous state, leading to abnormal function of the RNA polymerase and decreasing number of mature ribosomes. This could explain the quantitative reduction of TCOF1 mRNA observed only in the affected patient of Family 1 and not in the heterozygous asymptomatic parents and sister (Family 2 not tested).
To date, no clinical differences have been reported between the clinical phenotype associated with POLR1D (or POLR1C) mutations and the phenotype associated with TCOF1 mutations.5 The limited number of POLR1D-mutated patients with autosomal recessive mode of inheritance is too small to support any definitive conclusions; nevertheless, the craniofacial malformations of the patients in these two families were very typical of TCS and no additional symptoms were identified. The same high degree of inter- and intrafamilial variation as that classically described in TCS was also observed here: the girls in Family 1 and Family 2 had moderate craniofacial malformations, whereas the brother in Family 2 had a more severe phenotype. Psychomotor delay and cognitive disabilities are uncommon in TCS; in the previous cohort with POLR1D mutations, only one patient had delayed motor development and none had cognitive deficiency.5 The affected girl in Family 1 had normal motor development and was very clever. In Family 2, psychomotor and cognitive development was normal among siblings (learning difficulties and behavior problems were mainly due to hearing loss).
In conclusion, POLR1D mutations may be associated with either autosomal dominant or autosomal recessive TCS, which is an uncommon situation among monogenic disorders. This must be taken into account when evaluating sporadic TCS patients, especially if TCOF1 analysis is negative.
Disclosure
The authors declare no conflict of interest.
References
Thomson A . Notice of several cases of malformation of the external ear, together with experiments on the state of hearing in such persons. Month J Med Sci 1846;7:420.
Berry GA . Note on a congenital defect colobomata of the lower lid. R Lond Ophthalmic Hosp Rep 1889;12:255.
Collins E . Cases with symmetrical congenital notches in the outer part of each lower lid and defective development of malar bones. Tr Ophth Sol United Kingdom 1900;20:190.
Gorlin R, Cohen MJ, Hennekam R . Syndromes of the Head and Neck, 4th edn. Oxford University Press: New York, 2001:790–798.
Dauwerse JG, Dixon J, Seland S, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 2011;43:20–2.
Rovin S, Dachi SF, Borenstein DB, Cotter WB . Mandibulofacial dysostosis, a familial study of five generations. J Pediatr 1964;65:215–21.
Splendore A, Jabs EW, Félix TM, Passos-Bueno MR . Parental origin of mutations in sporadic cases of Treacher Collins syndrome. Eur J Hum Genet 2003;11:718–22.
Jones KL, Smith DW, Harvey MA, Hall BD, Quan L . Older paternal age and fresh gene mutation: data on additional disorders. J Pediatr 1975;86:84–8.
Dixon J, Hovanes K, Shiang R, Dixon MJ . Sequence analysis, identification of evolutionary conserved motifs and expression analysis of murine tcof1 provide further evidence for a potential function for the gene and its human homologue, TCOF1. Hum Mol Genet 1997;6:727–37.
Masotti C, Ornelas CC, Splendore-Gordonos A, et al. Reduced transcription of TCOF1 in adult cells of Treacher Collins syndrome patients. BMC Med Genet 2009;10:136.
Dixon J, Jones NC, Sandell LL, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA 2006;103:13403–8.
Valdez BC, Henning D, So RB, Dixon J, Dixon MJ . The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA 2004;101:10709–14.
Larkin RM, Guilfoyle TJ . Reconstitution of yeast and Arabidopsis RNA polymerase alpha-like subunit heterodimers. J Biol Chem 1997;272:12824–30.
Acknowledgements
We thank the families patients and family membersfor their help and support. We thank Dr Riehm for her collaboration. This work was in part supported by the French Franceschetti - Treacher Collins association Coline, http://netcoline.org/, and by a grant from the German Ministry of Education and Research for the CRANIRARE to D.W. and D.R.L. (BMBF 01GH1211B).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Figure S1
(JPEG 576 kb)
Rights and permissions
About this article
Cite this article
Schaefer, E., Collet, C., Genevieve, D. et al. Autosomal recessive POLR1D mutation with decrease of TCOF1 mRNA is responsible for Treacher Collins syndrome. Genet Med 16, 720–724 (2014). https://doi.org/10.1038/gim.2014.12
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/gim.2014.12