Abstract
The common frog (Rana temporaria) has become a model species in the fields of ecology and evolutionary biology. However, lack of genomic resources has been limiting utility of this species for detailed evolutionary genetic studies. Using a set of 107 informative microsatellite markers genotyped in a large full-sib family (800 F1 offspring), we created the first linkage map for this species. This partial map—distributed over 15 linkage groups—has a total length of 1698.8 cM. In line with the fact that males are the heterogametic sex in this species and a reduction of recombination is expected, we observed a lower recombination rate in the males (map length: 1371.5 cM) as compared with females (2089.8 cM). Furthermore, three loci previously documented to be sex-linked (that is, carrying male-specific alleles) in adults from the wild mapped to the same linkage group. The linkage map described in this study is one of the densest ones available for amphibians. The discovery of a sex linkage group in Rana temporaria, as well as other regions with strongly reduced male recombination rates, should help to uncover the genetic underpinnings of the sex-determination system in this species. As the number of linkage groups found (n=15) is quite close to the actual number of chromosomes (n=13), the map should provide a useful resource for further evolutionary, ecological and conservation genetic work in this and other closely related species.
Similar content being viewed by others
Introduction
Amphibians are important model organisms in developmental, ecological and evolutionary studies (Cano et al., 2004; Burggren and Warburton, 2007; Smith and Voss, 2007; Laurila et al., 2008; Hellsten et al., 2010). In addition, their phylogenetic placement between fishes and terrestrial amniotes makes them especially useful for studies of character evolution in vertebrates (Smith and Voss, 2006). One particular aspect of vertebrate evolution to which amphibian models can make—and have already made (Sumida and Nishioka, 2000; Schmid and Steinlein, 2001; Eggert, 2004; Matsuba et al., 2008; Nakamura, 2009)—significant contributions is the evolution of sex-determination systems. Sex-determination systems vary greatly among vertebrates, and amphibian studies bear potential to reveal not only yet unrecognized diversity, but also conserved aspects characterizing the ancestral vertebrate condition (Smith and Voss, 2009). However, due to their large genome sizes and the difficulties in raising some species, there is a general lack of genomic resources for amphibians making it difficult to locate sex-determining loci and genomic regions containing genes of ecological relevance (Sumida and Nishioka, 2000; Smith and Voss, 2009). To date, there are only two amphibian model species (that is, Xenopus spp and Ambystoma spp) for which physical and genetic linkage maps are available (Table 1; Kochan et al., 2003; Smith et al., 2005; Smith and Voss, 2009; Hellsten et al., 2010).
Efficient genetic linkage mapping requires that large number of offspring can be genotyped. To this end, many amphibian species are well suited for this purpose because large numbers of offspring are produced making it possible to obtain accurate estimates of marker positions in the map. In particular, microsatellite-based linkage maps provide excellent tools to locate sex-determining loci in species lacking heteromorphic sex chromosomes (Peichel et al., 2004; Chistiakov et al., 2006). Microsatellite linkage maps may help in the identification of sex-determining loci by looking at specific features in the heterogametic sex, such as a strong reduction in recombination between markers linked to the sex-determination region in male as compared with female meioses (Charlesworth et al., 2005). Likewise, a consistent heterozygous status of males for unique alleles in a given region would indicate a potential sex-determination role of this region (Peichel et al., 2004; Chistiakov et al., 2006).
Sex-determination systems in amphibians are very complex and varied (Hayes, 1998; Eggert, 2004). Sex-linked loci may differ among closely related species, among populations of the same species and even among individuals within populations (Sumida and Nishioka, 2000). Even occurrence of both male and female heterogamety in the same species has been demonstrated (Nakamura, 2009). To further complicate the picture, environmental factors may also influence sex determination. Although female heterogamety has been suggested to be the ancestral state of sex determination in amphibians, the origins of the sex chromosomes and the sex-determining genes are still unknown (Uno et al., 2008b). In Ranidae, the details of sex-determination systems have been best studied in Japanese ‘brown frogs’ (Rana sp). In these species, males are usually the heterogametic sex, although in some species (for example, R. rugosa), both ZZ/ZW and XX/XY determination systems are known (Uno et al., 2008b). Multiple sex-determining loci have been found in these frogs, and their linkage relationships with neighboring loci are very unstable and vary dramatically between populations (Sumida and Nishioka, 2000).
The common frog (Rana temporaria) is one of the most widely distributed amphibians in Europe, and it has been extensively studied in different ecological and evolutionary biology contexts (Savage, 1961; Cano et al., 2004; Laugen et al., 2002, 2003a, 2005; Hettyey et al., 2005; Lindgren and Laurila, 2005; Laurila et al., 2008; Jönsson et al., 2009). Many traits display latitudinal divergence across the Scandinavian peninsula, and common garden experiments have shown that this differentiation is genetically based and driven by natural selection (Laurila et al., 2002; Palo et al., 2003; Laugen et al., 2003b). However, the progress in understanding the genetic architecture of these traits (for example, quantitative trait loci number, location, and size effects) has been hampered by the lack of genetic resources, such as linkage and physical maps. In this species, males are the heterogametic sex (XX/XY), albeit heteromorphism in sex chromosomes is very weak (Popov and Dimitrov, 1999). Recent studies suggest that males tend to have a smaller genome size than females (Matsuba and Merilä, 2006), and three microsatellite loci with varying degree of linkage to sex in Fennoscandian common frog populations have been found (Matsuba et al., 2008; Alho et al., 2010). Genome-wide linkage studies would allow measurement of potential differences in recombination rates between the sexes, as well identification of the number and location of sex-determining loci.
The aim of this study was to create the first genetic linkage map of the common frog to aid evolutionary genetic studies of this species. To this end, we took advantage of the large number of microsatellite markers available for this species (Matsuba and Merilä, 2009), and genotyped 800 F1 offspring from a cross between two divergent common-frog populations from Sweden. Apart from creating a resource useful for evolutionary biology and population genetics of this and related species, we were particularly interested to gain insight into the organization of the sex-determination system in the species by (i) establishing the location of markers known to be linked to sex in wild adults, (ii) finding non-recombining regions in males, and (iii) estimating recombination rate differences between sexes.
Materials and methods
Frog sampling and husbandry
We collected one adult male Rana temporaria in a pond outside Lund (55° 43′ N, 13° 15′ E) on 3 April 2008, and one adult female near Kiruna (Jukkasjärvi, 67° 51′ N, 21° 02′ E) on 25 May 2008. Both the individuals were immediately transported to the laboratory in Uppsala. The male was kept in the laboratory at 4 °C in a plastic box filled with moist peat moss (Sphagnum sp.) until it was crossed with the female on May 27 to produce one full-sib family (see Räsänen et al., 2003 for crossing methods).
The eggs were raised in the laboratory (16 °C, 18-h-light:6-h-dark photoperiod) in four 3 l vials (ca. 250 eggs per vial) filled with reconstituted soft water (APHA, 1985) until they had hatched and reached stage 25 (that is, gills fully absorbed; Gosner, 1960). The water in the vials was changed completely every three days. The larval common garden experiment was started on 8 June 2009. 800 seemingly healthy tadpoles were placed individually in 1 l plastic vials filled with 0.8 l reconstituted soft water. Half of the tadpoles were raised in a walk-in temperature-controlled room set to 16 °C and the other half in a similar room set to 22 °C to check for genotype by environment interactions in follow-up quantitative trait locus mapping studies (Cano et al., in preparation). Photoperiod in both rooms was set to 18 h light:6 h dark. The tadpoles were fed ad libitum chopped spinach and the water was changed every 3 days. When the tadpoles approached metamorphosis (stage 42; occurrence of at least one forelimb; Gosner, 1960), the containers were checked daily. Metamorphosed tadpoles were sacrificed using MS-222 and preserved in 70% ethanol. Animal collection permits were granted by the Skåne and Norrbotten county boards, and the breeding experiment was conducted with a license from the Committee for Animal Experiments in Uppsala County.
Genotyping
Both parents and the F1 full-sib offspring were genotyped for a total of 113 polymorphic microsatellite loci, including 106 loci from Matsuba and Merilä (2009) and seven markers previously developed in other frog species (see Supplementary File 1). Detailed information of all these loci including their names, primer sequences, Genbank accession numbers, repeat motifs and informative meioses is available in Supplementary File 1. DNA was extracted from muscle tissue as described in Duan and Fuerst (2001). PCR conditions were optimized as follows: total volume of 10 μl consisted of 1xPhusion Flash High-Fidelity PCR Master Mix (F-548 Finnzymes, Vantaa, Finland), 0.5 μM of both forward (labeled with fluorochrome FAM, TET, or HEX) and reverse primers together with 1:10 diluted sample. All PCRs were multiplexed by combining six primer pairs in each reaction according to size and fluorochrome. Primer pairs with HEX-labeled forwards were diluted into final concentration of 0.3 μM and others into 0.2 μM; template DNA was diluted 1:5. PCRs were conducted in Piko Thermal Cycler (Finnzymes) using the following thermal profile: 98 °C for 1 min, 36 cycles of 98 °C for 1 s, 60 °C for 30 s, 72 °C for 30 s and final extension at 72 °C for 1 min. PCR products were then run (1:100 dilution) together with ET-ROX 550-size standard in MegaBACE1000 (GE Healthcare Life Sciences, Little Chalfont, UK) capillary electrophoresis instrument and genotypes were determined using the program Fragment Profiler 1.2 (GE Healthcare).
All the genotypes were double-read independently by two people. Less than 5% of the genotype readings were found to be inconsistent between the readers, and incompatible genotypes were blanked. Additional independent PCRs for these individual-locus combinations were carried out for further verification. These blanks were refilled with the correct genotypes if they worked out after the additional PCRs. Mendelian inheritance was checked with PedCheck software (O’Connell and Weeks, 1998) and using the ‘prepare’ option in CRIMAP program version 2.4 (Green et al., 1990). Inconsistent genotypes were re-run. If persistent, inconsistencies were removed from the dataset. Genotypes that were highly improbable due to low allele frequencies in the pedigree were eliminated (Jaari et al., 2009).
Linkage analysis
Linkage analyses were performed with CRIMAP 2.4 (Green et al., 1990) to obtain sex-average and sex-specific linkage genetic distances (that is, based on the parental meioses) for each linkage group. Initially, all markers were tested against each other with the ‘twopoint’ option and markers that clustered together with significant logarithm of odds (LOD) score support (>3.0) were treated as linkage groups. Framework maps were constructed with the ‘build’ option, and the best position of all markers within an ordered linkage group was then estimated with recurrent runs of the option ‘flips4’, until no better order could be found (best order map). Relative mapping distance was further tested within group using the ‘fixed’ option. All map distances are expressed as Kosambi centiMorgans (cM).
Cross-species homologies
To assess the degree of shared synteny with other amphibian species microsatellite, clone sequences were compared with the Xenopus tropicalis genome draft (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=8364) and Ambystoma t. tigrinum and A. t. mexicanum expressed sequence tag (EST) assemblies (http://salamander.uky.edu/ESTdb/blast.php) in cross-species BLAST searches using the program BLASTN 2.2.24+ (Zhang et al., 2000). Searches were done with the default conditions, except that the expectation value was set to a more stringent setting of 1e−5. A locus was considered to give a significant hit to the Xenopus and Ambystoma genomes if it provided a unique match at 1e−10 or lower (Dawson et al., 2006). If a locus did not provide a single unique hit but provided multiple matches, then it was unassigned unless the best hit had an expectation value at least 10 decimal places lower than the next best hit. Repeat motifs were masked using the low-complexity filter.
Results
Map size and linkage groups
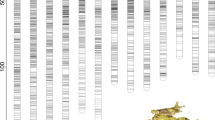
From a starting set of 113 amplifying microsatellite markers, six (Bfg005, Bfg014, Bfg038, Bfg084, Bfg141 and Bfg252) were not informative (that is, both parents were homozygous). Additional three markers (Bfg115, Bfg147 and Bfg211) were informative but did not show significant linkage with any other marker. The remaining 104 markers were linked and formed 15 linkage groups (Figure 1). The male parent was informative (that is, heterozygous) for 81.4% of the markers and the female parent for 76.9%. Among the 15 linkage groups found, 10 had less than 5 markers and 5 were triplets or duplets which may be in reality linked to other groups (R. temporaria karyotype consisting of 13 chromosome pairs; Popov and Dimitrov, 1999). The sex-average map length was 1698.8 cM (Figure 1), and sex-specific maps of females and males were 2089.8 and 1371.5 cM (Figure 2), respectively. The large size of F1 family gave high power to estimate significant linkage (that is, all linked loci had LOD >10) that resulted in a total lack of ambiguous locations. The map reported here is, therefore, a framework map, and running the ‘flips’ option in CRIMAP did not alter the order of the markers as obtained by the ‘build’ option. Recombination rate in females was 1.8 times larger than in males (average, 95% confidence interval; females 0.37, 0.36–0.39; males 0.21, 0.19–0.23). Contrasting recombination rates between sexes were only found in five of the linkage groups (LGs: 2, 3, 7, 9 and 11; see Figure 2).
Sex-specific linkage maps in Kosambi cM (that is, for each group female meioses are on the left and male meioses on the right). Markers previously found linked to sex are marked with an asterisk (Matsuba et al., 2008, Alho et al., 2010) or underlined (Matsuba unpublished). Grey lines show contrasting recombination distances between sexes.
Sex-linked markers
An interesting feature of this map is that six markers known to be linked to phenotypic sex in wild individuals in Northern Finland and Sweden (that is, Bfg201, Bf266 and RtSBO3 [Matsuba et al., 2008; Alho et al., 2010] and Bfg053, Bfg093 and Bfg191 [Matsuba, unpublished]) mapped to the same linkage group. Marker Bfg028, which was sex-linked in a population from Southern Sweden (Tvedöra 55°42′N, 13°26′E; Matsuba, unpublished), mapped to a different linkage group (that is, LG7).
Shared synteny
Both the default and relaxed BLASTN settings (Dawson et al., 2006) yielded the same results. Eight markers (Bfg 195, Bfg191, Bfg172, Bfg051, Bfg131, Bfg072, Bfg028 and Bfg008) produced significant matches with unplaced X. tropicalis scaffolds. Two significant matches (Bfg131 and Bfg072) were obtained with Ambystoma t. tigrinum unmapped contigs. Therefore, no proper assessment of shared synteny was possible.
Discussion
This is a first generation linkage map for R. temporaria, and in addition to providing an important resource for further evolutionary, ecological and conservation genetic studies, it may advance our understanding of the mechanisms behind genetic sex determination in amphibians by helping to locate sex determination gene/s in anurans in which the genetic basis of sex determination is poorly understood (Sumida and Nishioka, 2000; Schmid and Steinlein, 2001; Matsuba and Merilä, 2009; but see Uno et al., 2008a, 2008b).
Genetic sexual determination
Previous studies have reported male-specific alleles in markers Bfg201, Bf266 and RtSBO3 in the common frog (Matsuba et al., 2008; Alho et al., 2010). The sex linkage was observed in Northern Fennoscandian populations, but not in Southern Fennoscandian populations (Matsuba et al., 2008). Our results show that these three markers belong to the same linkage group (LG2), suggesting that the sex-determining locus/loci is/are located in LG2. The contention that LG2 carries the sex-determining gene is also supported by two additional lines of evidence.
First, recombination rate in male meioses was dramatically lower as compared with female meioses (Figure 2), as expected in the heterogametic sex (Rice, 1987). Second, all the markers were heterozygous in LG2 in the male parent, whereas in the female parent, only 7 out of 14 were heterozygous or informative, in agreement with the expectation of a predominantly heterozygous state in the sex-specific chromosome (Charlesworth et al., 2005). However, the recombination distances among these three sex-linked markers were fairly large, indicating that some of them may not be tightly linked to the sex-determining locus in our male parent. Given that our male parent originates from a population in Southern Sweden, the large recombination rate among these markers would explain why no significant linkage with adult phenotypic sex was found in the wild (Matsuba et al., 2008). The other three markers belonging to LG2 (Bfg053, Bfg093 and Bfg191) had significant linkage disequilibrium with Bfg201 and had male-specific alleles in the wild adult individuals of a Northern Finnish population (Kilpisjärvi 69°04′N, 20°46′E; Matsuba, unpublished) further strengthening the interpretation that LG2 is linked to sex determination.
Our results also revealed other linkage groups (that is, LG3, 7 and 9) that show extreme male-specific reduction in recombination rates (Figure 2), which is consistent also with the possibility of several sex-determining loci located in different linkage groups (Wright and Richards, 1993). For instance, Bfg028 is located in LG7 and was linked to sex in adults from a population of Southern Sweden (Tvedöra 55°42′N, 13°26′E; Matsuba unpublished). Therefore, although this map finds strong evidence of LG2 being a sex-linkage group, it also provides new candidate regions to be tested for linkage with sex determination.
Mapping and population/conservation genetic prospects
Despite the sparse coverage (that is, 108 markers in a ∼4.2 Mb genome) of the map, we found 15 linkage groups that were quite close to the actual number of chromosomes in this species (n=13; Popov and Dimitrov, 1999; Peskov et al., 2004). When ordered according to their size, the linkage groups display a clear decreasing pattern in their length as expected from the decreasing physical size observed in cytological studies (Peskov et al., 2004). Furthermore, only five groups were triplets or duplets and may belong to other linkage groups, but we didn’t have enough resolution to assign them. This map is, therefore, a good starting point with enough resolution for mapping quantitative trait loci of traits that exhibit adaptive differentiation among the populations used in the cross, such as developmental time and size at metamorphosis (Laurila et al., 2002; Palo et al., 2003; Laugen et al., 2003a, 2003b). It also allows selection of sets of unlinked markers—or markers in different linkage groups—for studies of neutral genetic processes using microsatellites (Selkoe and Toonen, 2006). Likewise, studies wishing to investigate sex-specific migration rates, sex-specific survival rates of embryos or larval stages—or primary sex ratios—will benefit directly from our results.
Caveats and future directions
A denser coverage would be needed in order to determine whether the smallest linkage groups correspond to different chromosomes or are fragments of other chromosomes. Big gaps in some of the linkage groups may make it difficult to locate quantitative trait loci and sex-determining regions. However, the coverage can be increased in the future, as high-throughput sequencing methods start allowing rapid and cost-effective development of genomic resources even for non-model species (Wheat, 2010). Targeting functionally relevant genes by placing markers within or close to genes of known functions (Shikano et al., 2010) would also facilitate a proper assessment of the degree of synteny among amphibian genomes and other taxa.
Conclusions
We have described the first linkage map for the common frog. This map is the densest available for species in the family Ranidae, and one of the most complete ones in amphibians in general (Table 1). With the exception of the genome draft for X. tropicalis (Hellsten et al., 2010), only a map for Ambystoma spp (Smith et al., 2005) provides similar coverage relative to its genome size. Of particular interest in our study is also the finding of a linkage group carrying several sex-linked markers. This can serve as a starting point of further studies of genetic sex determination in this and related species. Likewise, given the lack of genomic resources for most amphibian species, the map described in this paper should provide a valuable resource for future comparative genomic studies of amphibians and mapping of ecologically relevant traits in the common frog.
References
Alho JS, Matsuba C, Merilä J (2010). Sex reversal and primary sex ratios in the common frog (Rana temporaria). Mol Ecol 19: 1763–1773.
APHA (American Public Health Association) (1985). Standard Methods for the Examination of Water and Waste Water, 14th edn. APHA, AWWA, WPCF: Washington DC, USA, 1448 pp.
Berset-Brändli L, Jaquiéry J, Broquet T, Ulrich Y, Perrin N (2008). Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc R Soc B 1642: 1577–1585.
Burggren WW, Warburton S (2007). Amphibians as animal models for laboratory research in physiology. ILAR J 48: 260–269.
Cano JM, Laurila A, Palo JU, Merilä J (2004). Population differentiation in G matrix structure due to natural selection in Rana temporaria. Evolution 58: 2013–2020.
Charlesworth D, Charlesworth B, Marais G (2005). Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128.
Chistiakov DA, Helleman B, Volckaert FAM (2006). Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture 255: 1–29.
Dawson DA, Burke T, Hansson B, Pandhal J, Hale MC, Hinten GN et al. (2006). A predicted microsatellite map of the passerine genome based on chicken-passerine sequence similarity. Mol Ecol 15: 1299–1320.
Duan W, Fuerst PA (2001). Isolation of a sex-linked DNA sequence in cranes. J Hered 92: 392–397.
Eggert C (2004). Sex determination: the amphibian models. Reprod Nutr Dev 44: 539–549.
Gosner KL (1960). A simplified table for staging anuran embryos and larvae. Herpetologica 6: 183–190.
Graf JD (1993) In: O’Brien SJ (ed). Genetic Maps: Locus Maps of Complex Genomes. Cold Spring Harbor Laboratory Press: New York, pp 4330–4331.
Graf JD (1989a). Genetic mapping in Xenopus laevis: eight linkage groups established. Genetics 123: 389–398.
Graf JD (1989b). Sex linkage of malic enzyme in Xenopus laevis. Experientia 45: 194–196.
Green P, Falls K, Crook S (1990). Documentation for CRIMAP, Version 2.4. Washington University School of Medicine: St Louis.
Hayes TB (1998). Sex determination and primary sex differentiation in Amphibians: genetic and developmental mechanisms. J Exp Zool 281: 373–399.
Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V et al. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636.
Hettyey A, Laurila A, Herczeg G, Jönsson KI, Kovács T, Merilä J (2005). Does testis weight decline towards the Subarctic? A case study on the common frog, Rana temporaria. Naturwissenschaften 92: 188–192.
Hotz H, Uzzell T, Berger L (1997). Linkage groups of protein-coding genes in western palearctic water frogs reveal extensive evolutionary conservation. Genetics 147: 255–270.
Jaari S, Li MH, Merilä J (2009). A first-generation microsatellite-based genetic linkage map of the Siberian jay (Perisoreus infaustus): insights into avian genome evolution. BMC Genom 10: 1.
Jönsson KI, Herczeg G, O’Hara RB, Söderman F, ter Schure AFH, Larsson P et al. (2009). Sexual patterns of pre breeding energy reserves in the common frog Rana temporaria along a latitudinal gradient. Ecography 32: 831–839.
Kochan KJ, Wright DA, Schroeder LJ, Shen J, Morizot DC (2003). Genetic linkage maps of the West African clawed frog Xenopus tropicalis. Dev Dyn 226: 99–102.
Laugen AT, Laurila A, Jönsson KI, Söderman F, Merilä J (2005). Do common frogs (Rana temporaria) follow Bergmann's rule? Evol Ecol Res 7: 717–731.
Laugen AT, Laurila A, Merilä J (2002). Maternal and genetic contributions to geographical variation in Rana temporaria larval life-history traits. Biol J Linn Soc 76: 61–70.
Laugen AT, Laurila A, Merilä J (2003a). Latitudinal and temperature-dependent variation in embryonic development and growth in Rana temporaria. Oecologia 135: 548–554.
Laugen AT, Laurila A, Räsänen K, Merilä J (2003b). Latitudinal countergradient variation in the common frog (Rana temporaria) developmental rates—evidence for local adaptation. J Evol Biol 16: 996–1005.
Laurila A, Karttunen S, Merilä J (2002). Adaptive phenotypic plasticity and genetics of larval life histories in two Rana temporaria populations. Evolution 56: 617–627.
Laurila A, Lindgren B, Laugen AT (2008). Antipredator defenses along a latitudinal gradient in Rana temporaria. Ecology 89: 1399–1413.
Lindgren B, Laurila A (2005). Proximate causes of adaptive growth rates: growth efficiency variation among latitudinal populations of Rana temporaria. J Evol Biol 18: 820–828.
Matsuba C, Merilä J (2006). Genome size variation in the common frog Rana temporaria. Hereditas 143: 155–158.
Matsuba C, Merilä J (2009). Isolation and characterization of 145 polymorphic microsatellite loci for the common frog (Rana temporaria). Mol Ecol Res 9: 555–562.
Matsuba C, Miura I, Merilä J (2008). Disentangling genetic vs environmental causes of sex determination in the common frog, Rana temporaria. BMC Genet 3: 8.
Nakamura M (2009). Sex determination in amphibians. Semin Cell Dev Biol 20: 271–282.
Nishioka M, Ohtani H (1986). Detection of chromosomes bearing the loci for blue and olive mutations in Rana nigromaculata. Sci Rep Lab Amphibian Biol Hiroshima Univ 8: 1–27.
Nishioka M, Ohtani H, Sumida M (1980). Detection of chromosomes bearing the loci for seven kinds of proteins in Japanese pond frogs. Sci Rep Lab Amphibian Biol Hiroshima Univ 4: 127–184.
Nishioka M, Ohtani H, Sumida M (1987). Chromosomes and the sites of five albino gene loci in the Rana nigromaculata group. Sci Rep Lab Amphibian Biol Hiroshima Univ 9: 1–52.
Nishioka M, Sumida M (1994a). The position of sex-determining genes in the chromosomes of Rana nigromaculata and Rana brevioda. Sci Rep Lab Amphibian Biol Hiroshima Univ 13: 51–97.
Nishioka M, Sumida M (1994b). The differences in recombination rate between the male and female in Rana nigromaculata and Rana brevipoda. Sci Rep Lab Amphibian Biol Hiroshima Univ 13: 99–136.
Nürnberger B, Hofman S, Förg-Brey B, Praetzel G, Maclean A, Szymura JM et al. (2003). A linkage map for the hybridising toads Bombina bombina and B. variegata (Anura: Discoglossidae). Heredity 91: 136–142.
O’Connell JR, Weeks DE (1998). PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63: 259–266.
Palo JU, O’Hara RB, Laugen AT, Laurila A, Primmer CR, Merilä J (2003). Latitudinal divergence of common frog (Rana temporaria) life history traits by natural selection: evidence from a comparison of molecular and quantitative genetic data. Mol Ecol 12: 1963–1968.
Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J et al. (2004). The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol 14: 1416–1424.
Peskov VN, Kotserzhynska IM, Manilo VV, Pisanets EM (2004). Morphological differentiation and diagnostic of brown frogs Rana arvalis, R. temporaria and R. dalmatina (Amphibia, Ranidae) from the territory of Ukraine. Vestnik Zoologii 38: 29–40.
Popov P, Dimitrov B (1999). Karyotype study of Rana camerani and comparisons with the other 26-chromosome European brown frog species (Amphibia, Anura). Cytobios 97: 13–22.
Räsänen K, Laurila A, Merilä J (2003). Geographic variation in acid stress tolerance of the moor frog, Rana arvalis. I. Local adaptation. Evolution 57: 352–362.
Rice WR (1987). The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914.
Savage RM (1961). Ecology and Life-history of the Common Frog. Sir Isaac Pitman & Sons: London.
Schmid M, Steinlein C (2001). Sex chromosomes, sex-linked genes, and sex determination in the vertebrate class Amphibia. EXS 91: 143–176.
Selkoe KA, Toonen RJ (2006). Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9: 615–629.
Shikano T, Ramadevi J, Merilä J (2010). Identification of local and habitat dependent selection: Scanning functionally important genes in nine-spined sticklebacks (Pungitus pungitus). Mol Biol Evol 27: 2775–2789.
Smith JJ, Kump DK, Walker JA, Parichy DM, Voss SR (2005). A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: enabling gene mapping and comparative genomics in Ambystoma. Genetics 171: 1161–1171.
Smith JJ, Voss SR (2006). Gene order data from a model amphibian (Ambystoma): new perspectives on vertebrate genome structure and evolution. BMC Genomics 7: 219.
Smith JJ, Voss SR (2007). Bird and mammal sex-chromosome orthologs map to the same autosomal region in a salamander (Ambystoma). Genetics 177: 607–613.
Smith JJ, Voss SR (2009). Amphibian sex determination: segregation and linkage analysis using members of the tiger salamander species complex (Ambystoma mexicanum and A. t. tigrinum). Heredity 102: 542–548.
Sumida M (1996). Inheritance and linkage analysis of ten enzyme and blood protein loci in the Japanese brown frog Rana japonica. Biochem Genet 34: 375–388.
Sumida M, Nishioka M (1994a). Sci Rep Lab Amphibian Biol Hiroshima Univ 13: 173–195.
Sumida M, Nishioka M (1994b). A pronounced sex difference when two linked loci of the Japanese brown frog Rana japonica are recombined. Biochem Genet 32: 361–369.
Sumida M, Nishioka M (2000). Sex-linked genes and linkage maps in amphibians. Comp Biochem Physiol B Biochem Mol Biol 126: 257–270.
Uno Y, Nishida C, Oshima Y, Yokoyama S, Miura I, Matsuda Y et al. (2008a). Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosome Res 16: 637–647.
Uno Y, Nishida C, Yoshimoto S, Ito M, Oshima Y, Yokoyama S et al. (2008b). Diversity in the origins of sex chromosomes in anurans inferred from comparative mapping of sexual differentiation genes for three species of the Raninae and Xenopodinae. Chromosome Res 16: 999–1011.
Wheat CW (2010). Rapidly developing functional genomics in ecological model systems via 454 transcriptome sequencing. Genetica 138: 433–451.
Wright DA, Richards CM (1983). Two sex-linked loci in the leopard frog, Rana pipiens. Genetics 103: 249–261.
Wright DA, Richards CM (1993). Linkage groups in the leopard frog, Rana pipiens. In: O’Brien SJ (ed). Genetic Maps: Locus Maps of Complex Genomes. Cold Spring Harbor Laboratory Press: New York, pp 4326–4329.
Wright DA, Richards CM, Nace GW (1980). Inheritance of enzymes and blood proteins in the leopard frog, Rana pipiens: three linkage groups established. Biochem Genet 18: 591–616.
Wright DA, Richards CM, Nace GW (1983). Inheritance of enzymes and blood proteins in the leopard frog, Rana pipiens: three linkage groups established. Curr Top Biol Med Res 10: 287–311.
Zhang Z, Schwartz S, Wagner L, Miller W (2000). A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–221.
Acknowledgements
We thank Jarmo Saarikivi and Jon Loman for help in obtaining individuals for parental generation, Carin Andersson for help in rearing the tadpoles. Marika Karjalainen and Kirsi Kähkönen helped with laboratory work. Thanks are also due to Nicolas Perrin, Chikako Matsuba and Jussi Alho for discussions and feedback at various stages of this work. John Loehr revised the English. Our research was supported by grants from Swedish Research Council (AL), Academy of Finland (JM and JMC) and Ministry of Education, Finland (JM).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Cano, J., Li, MH., Laurila, A. et al. First-generation linkage map for the common frog Rana temporaria reveals sex-linkage group. Heredity 107, 530–536 (2011). https://doi.org/10.1038/hdy.2011.39
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/hdy.2011.39
Keywords
This article is cited by
-
A high-density genetic map construction and sex-related loci identification in Chinese Giant salamander
BMC Genomics (2021)
-
The genetic contribution to sex determination and number of sex chromosomes vary among populations of common frogs (Rana temporaria)
Heredity (2016)
-
High-density sex-specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex
Heredity (2016)