Abstract
Many organisms display codispersal of offspring, but fewer display codispersal of compatible gametes. This mechanism enhances the ability of a species to colonize after long distance dispersal as a mechanism of reproductive assurance, but it also fosters inbreeding and potential reduction in fitness. Here we investigated both long distance dispersal and inbreeding in the bird’s nest fungus Cyathus stercoreus, a dung and mulch-associated fungus with a splash cup fruiting body appearing like a miniature bird’s nest of ‘eggs‘ or peridioles that contain thousands of mating compatible meiotic spores. To investigate the genetic structure in the species, six North American urban populations were hierarchically sampled and genotyped using 10 microsatellite markers. We detected significant levels of inbreeding through heterozygote deficiencies at four loci, with global FIS=0.061. Dispersal limitation was suggested by both spatial autocorrelation and the detection of population structure between Louisiana and Michigan using clustering and F-statistics. Although inbreeding may facilitate colonization by the fungus, it has a negative effect on the fitness of populations as estimated from a 15% reduction in growth rates of inbred strains relative to outcrossed. Mating tests revealed that C. stercoreus has a higher estimated number of mating-type alleles (MAT-A= 39, MAT-B= 24) than other species of bird’s nest fungi, which would increase its outcrossing efficiency. We speculate that the increased number of mating-type alleles is the result of a recent range and population size expansion into urban environments.
Similar content being viewed by others
Introduction
Inbreeding is broadly defined as mating between related individuals occurring at a rate higher than random mating. Inbreeding increases homozygosity within a population and can cause an overall fitness decline, referred to as inbreeding depression (Keller and Waller, 2002). Increased homozygosity causes inbreeding depression by reducing potential heterosis and by forming homozygous genotypes at loci with deleterious recessive or partly recessive mutations (Charlesworth and Charlesworth, 1999; Charlesworth and Willis, 2009). Inbreeding avoidance manifests itself in many forms that are typically considered facets of an organism’s mating system, including behavioral mechanisms, genetic systems such as self-incompatibility loci, as well as dispersal mechanisms that decrease the likelihood of consanguineous mating.
Although inbreeding depression has been demonstrated numerous times among animals and plants (Keller and Waller, 2002; Charlesworth and Willis, 2009), there have been very few studies that assess if similar patterns can be found among fungal populations. Most fungal populations are predicted to be largely outcrossing because of the high potential for long distance wind dispersal of microscopic propagules (Ingold, 1953; Lacey, 1996; James et al., 1999). Inbreeding in fungi can occur through three distinct modes: haploid selfing, diploid selfing and matings among relatives (Billiard et al., 2012). Only homothallic fungi are capable of haploid selfing wherein mating occurs among fungal gametes that are genetically identical. In contrast, diploid selfing occurs between independent gametes of the same diploid individual and is similar to that of selfing in flowering plants. All forms of fungal inbreeding serve to reduce heterozygosity and may lead to inbreeding depression. Most fungi are considered heterothallic and are unable to undergo haploid selfing, and in this paper the discussion of inbreeding refers specifically to diploid selfing and consanguineous matings.
Both mating systems and the genetic structure of fungal populations have been widely studied, but few studies have linked the two. An explanation of fungal mating systems as mechanisms for inbreeding avoidance would be informed by more studies on whether inbreeding depression occurs in natural populations of fungi. Inbreeding depression can be readily measured in natural populations, but little effort has been made to test for it in fungi, perhaps because a free-living haploid phase should eliminate many recessive deleterious alleles, the leading hypothesized basis of inbreeding depression (Charlesworth and Willis, 2009). However, fungi could express inbreeding depression through recessive mutations expressed in the diplophase such as those governing sexual characteristics. These studies are supported by investigations of inbreeding depression in diplophase characteristics (such as fruiting body production) in both natural as well as cultivated fungal species (Eugenio and Anderson, 1968; Leslie and Raju, 1985; Xu, 1995). This study is an initial step toward bridging the gap between population structure in natural fungal populations and the dispersal mechanisms and mating systems that influence the structure.
The fungal kingdom harbors an immense diversity of species that exhibit a large variety of mating systems and dispersal mechanisms. Among this diversity, one family of fungi, the Nidulariaceae (Agaricomycetes: Basidiomycota), commonly known as the ‘bird’s nest fungi’, stand out as having one of the most unique dispersal mechanisms, which may facilitate inbreeding. Nidulariaceae ‘package’ numerous spores into small egg-like peridioles produced in an inverted cone shaped fruiting body, the ‘nest’. The nest forms a splash cup that facilitates the ejection of the entire peridiole from the cup by drops of rain, and through a mass of adhesive hyphae (the hapteron), the peridioles cling to a surface after ejection. Through forcible launch, a single raindrop can launch an entire mass of thousands of spores up to 2.4 m (Brodie, 1975). Thus by packaging spores together, the force of a raindrop can be effectively transferred to a mass of spores in a manner that could not be achieved by any spore individually. The tradeoff is that a large number of spores derived from a single fruiting body are dispersed in close proximity, which is likely to facilitate inbreeding. Working to facilitate outcrossing, the sticking action of the hapteron allows peridioles to cling to vegetation for possible future ingestion and further dispersal by herbivores (Brodie, 1951). Evidence for this adaptation for herbivore dispersal can be seen with the species Cyathus stercoreus, by the fact it is primarily coprophilous and spores will not germinate until incubation at 40 °C for 48 h (Brodie, 1975). Although several species of Nidulariaceae are globally distributed, the expected mean dispersal distance of bird’s nest fungi is considered very small when compared with other aerial spore dispersing species.
Most species of the Nidulariaceae studied have a heterothallic, tetrapolar mating system, which prevents them from undergoing haploid selfing (Raper, 1966; Brodie, 1975). In the tetrapolar mating system, two independently inherited loci (MAT-A and MAT-B) control the mating type of a mycelium, a mechanism that may have evolved to reduce diploid selfing, because it reduces the compatibility during diploid selfing by one half. Compatible mating only occurs between homokaryotic mycelia (genetically haploid) that have different alleles at both the MAT-A and MAT-B mating-type loci (Brown and Casselton, 2001). Compatible homokaryons form a heterokaryotic mycelium by reciprocal nuclear exchange, resulting in a mycelium comprised of two nuclear genotypes that is genetically diploid. In Nidulariaceae, homokaryons have exactly one nucleus per cell (termed monokaryotic), and heterokaryons have typically two nuclei per cell (dikaryotic). Most members of the Agaricomycetes (mushroom-forming fungi) have dozens to hundreds of mating types per locus (Raper, 1966). These high numbers of mating types are maintained by symmetric overdominant selection and result in an extraordinarily high outbreeding efficiency—the probability that outcrossing among random haploid individuals will be compatible (Kües et al., 2011). In contrast, natural populations of the bird’s nest fungi have been reported to have a low number of mating types. In Cyathus striatus only four mating-type factors at the MAT-A locus and five mating-type factors at the MAT-B locus were found among 18 fruit bodies of C. striatus collected from Europe (Fries, 1940). Only 3 mating-type factors at the MAT-A locus and 11 at the MAT-B locus among 15 fruit bodies of Crucibulum laeve from Sweden were found (Fries, 1943). Theory suggests that the number of mating types that a species can maintain is directly related to its effective population size (Wright, 1939; Simchen, 1967). Thus, the bird’s nest fungi are predicted to have small population sizes, and this may be due to low dispersability or high variance in fecundity.
These observations of low dispersal distance, codispersal of sibling gametes and small effective population size predict that bird’s nest fungi are likely to display inbreeding in natural populations. Thus, bird’s nest fungi are an excellent system to investigate for evidence of inbreeding and inbreeding depression in mushroom fungi. In order to obtain a large enough sampling size for mating-type allele and microsatellite analysis, an aggressive species found in urban environments, C. stercoreus, was chosen as the study species. C. stercoreus may be commonly found on dung, but urbanization and modern landscaping techniques have created a vast amount of new habitat for it as well other species of Nidulariaceae. Mulched woodpiles in urban and rural environments seem to be ideal substrates for some species of Nidulariaceae. Other anthropogenically modified habitats often present substrates for colonization by Nidulariaceae, such as rotting fiber sacks or mats, nursery pots, and old boards (Brodie, 1975).
The main goals of this study were to determine the level of inbreeding occurring within populations of C. stercoreus and to determine the spatial structure of populations. For this purpose, deviation from Hardy–Weinberg equilibrium of two distinct populations (approximately 1500 km apart), each with three sub-populations, was estimated using microsatellite markers. We predicted that high levels of inbreeding are occurring at both the local and regional scale. As inbreeding reduces effective population size and thus the number of mating-type alleles that would be expected at equilibrium, we also estimated the number of mating-type alleles using crosses among monokaryons obtained from 18 C. stercoreus fruiting bodies. We predicted that the number of mating-type alleles would be similar to other species of bird’s nest fungi and significantly lower compared with species with large population sizes and random mating conditions. In addition, we sought to determine if C. stercoreus is experiencing inbreeding depression using growth rates of inbred and outcrossed specimens in the laboratory.
Materials and methods
Sample collection
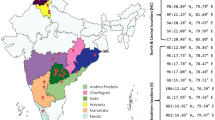
In the summer and fall of 2010, 29 specimens of C. stercoreus were collected from three different areas in Michigan. Michigan samples were primarily collected from mulch beds in urban areas near university campuses. During October of 2011, 43 samples were collected from mulched urban areas in three locations in Baton Rouge, LA, USA (Table 1). Once a patch of C. stercoreus was identified in decaying mulch, a small contiguous piece of mulch no larger than 2′ × 2′ and as small as two single fruiting bodies was collected. All the fruiting bodies in a small patch were assumed to be genetically identical. Once a sample was collected, no samples were collected from the same mulch bed unless another patch was located 10 m or more away from other collected samples within the bed.
Mating-type determination
Twelve Michigan specimens were selected (four from Grand Rapids, three from Ann Arbor and five from the Lansing population) for estimating numbers of mating types. To recover single-basidiospore isolates of each specimen, two to three peridioles were removed from a single fruiting body, lacerated and tweezed apart in order to dislodge spores from the capillitial tissue. The spore suspension was then passed through 40 μm nylon mesh and then placed in an incubator at 40 °C for at least 48 h. After incubation, 200–500 μl of the spore suspension was spread on a Petri dish containing modified Brodie medium and antibiotics (penicillin and streptomycin at 125 mg l−1). Modified Brodie medium contains 5.0 g of malt extract, instead of 5.0 g of maltose (Brodie, 1975). After spores were allowed to germinate for approximately 24 h, individual germlings were transferred to Petri dishes containing modified Brodie medium. Monokaryotic tester strains were isolated from dikaryotic cultures obtained from the Centraalbureau voor Schimmelcultures (Taiwan (CBS 337.81), Edmonton, Canada (CBS 378.80), the United Kingdom (CBS 534.97)) by fruiting the dikaryons according to the protocol of Flegler (1979).
From each specimen, 10–12 individual monokaryons were selected and a half-diallel cross was performed. The results were used to assign each of the four possible tetrapolar mating types to the monokaryotic cultures. In several cases, only 1–3 mating types were recovered from a specimen. One of each identified mating type from each specimen was then used in a 40 × 40 half-diallel cross. Successful mating was determined by the presence of clamp connections at the margin of the mycelia, the characteristic structure involved in maintaining the binucleate condition of the dikaryon. In numerous cases, the pairings were subcultured in order to observe dikaryotic hyphae for the presence of clamp connections because the clamps were absent at the margin of the colonies. Failure to form a dikaryon is indicative of a shared mating-type allele. Distinguishing incompatible pairings as the result of a shared MAT-A allele or a shared MAT-B allele utilized morphological examination of the pairings. Incompatibility because of a shared MAT-A allele was assigned to pairings that resulted in the emergence of sickly looking hyphae with many short branches. Common MAT-A reactions occur because of formation of heterokaryons that are not able to undergo proper conjugate division (Raper and Raper, 1968). Incompatibility because of a common MAT-B allele, in which reciprocal nuclear migration has failed to occur, was assigned to pairings that resulted in a distinct zone of inhibition in the interaction zone and two distinctive hyphal growth types on subculturing. Mating-type alleles were assigned and number of mating types at each locus was estimated by solving, by numerical approximation implemented in Mathematica 7 (Champaign, IL, USA), the maximum likelihood expression of (O’Donnell and Lawrence, 1984): N(m-1)(N−n)−(N−2)m=0, where N=the maximum likelihood estimate of allele number, m=number of sampled dikaryons and n=number of mating-type alleles observed. Only dikaryons in which both mating types at a particular MAT locus had been recovered were used for this calculation.
Genome sequencing
The monokaryotic strain CS4.14 was used for whole genome sequencing using the Illumina sequencing platform (Illumina, San Diego, CA, USA). The strain was cultivated in 500 ml of liquid modified Brodie medium and harvested by filtration after 11 days of growth. The mycelium was freeze dried, and DNA extraction was conducted using a standard hexadecyltrimethylammonium bromide isolation method for filamentous fungi as previously described (James et al., 2008). The genomic DNA was used to create a sequencing library of fragments of mean size 650 bp (fractionated by agarose gel electrophoresis) using NEBNext Reagents for DNA Sample Preparation for Illumina (New England BioLabs, Ipswich, MA, USA). The library was barcoded and run on 1/3 of a lane of an Illumina GAIIx analyzer at the University of Michigan DNA Sequencing Core, using paired end reads of the fragments run to 108-bp cycles. This resulted in 30.1 × 106 reads, which were assembled into scaffolds using Velvet 1.1.04 (Zerbino and Birney, 2008) with k-mer length 49 and automatic cutoff for read depth. This lead to an assembly of total size 71.07 Mbp, an assembly N50 of 34 194 bp, and a total of 9105 contigs of size >200 bp.
Microsatellite analysis
The assembled genome sequence was then used to develop microsatellite markers using the script Misa (Thiel et al., 2003) to identify simple sequence repeats and the software Primer3 (Rozen and Skaletsky, 2000) to design primers. We tested 45 potential microsatellite markers, 30 containing the longest dinucleotide repeats and 15 containing the longest tri and hexa nucleotide repeats by screening for amplification and polymorphism among 5 to 11 strains. Markers displaying consistent amplification and interpretable patterns were then used to assess diversity in our samples (Table 1). Samples were prepared by DNA extraction from a single fruiting body of each collected specimen. The peridioles were removed from the fruiting body and the remaining tissue was used for hexadecyltrimethylammonium bromide DNA extraction. Microsatellite genotypes were determined using methods (Schuelke, 2000) that reduce the expense of labeled primers by using an M13 5′-overhang on the locus-specific forward primer. The PCR reaction consists of locus-specific forward and reverse primers and a fluorescently labeled M13 primer in the same reaction. PCR was performed using ExTaq proofreading DNA polymerase (TaKara, Otsu, Japan) with 12.5 nM forward primer, 12.5 nM 6-FAM labeled M13F primer, 416 nM reverse primer in a 6 μl reaction. PCR amplifications conditions were: 94 °C (5 min), then 30 cycles at 94 °C (30 s)/56 °C (45 s)/72 °C (45 s), followed by 8 cycles 94 °C (30 s)/53 °C (45 s)/72 °C (45 s), and a final extension at 72 °C for 10 min (Schuelke, 2000). One microliter of the PCR product was then added to 12 μl formamide and 0.1 μl ROX GS500 standard (Applied Biosystems, Foster City, CA, USA) and run on an ABI 3730 capillary sequencer at the University of Michigan Sequencing Core. Fragment lengths were determined using GeneMarker software (SoftGenetics, State College, PA, USA). Loci were checked for the presence of null alleles and other possible genotyping errors using MICRO-CHECKER (Van Oosterhout et al., 2004).
Estimating inbreeding depression
To investigate if C. stercoreus displays inbreeding depression, the growth rates of inbred and outbred genotypes were compared using the dikaryons generated in the crossing experiment. Although only one measurement of fitness, growth rate has been widely used as an indicator of fitness because it is easily observable and has been correlated other fitness components in different species of fungi, such as spore production and head-to-head genotype competition (deVisser et al., 1997; Pringle and Taylor, 2002; Schoustra et al., 2009). Growth rate was shown by Xu (1995) to demonstrate inbreeding depression through slower growth rates of inbred Agaricus bisporus heterokaryons relative to outcrossed. The fitness of 18 inbred dikaryotic genotypes were estimated by constructing dikaryons through mating between monokaryotic individuals obtained from the same fruiting body (diploid selfing). The fitness of 27 outbred genotypes were estimated from matings between monokaryons isolated from different fruiting bodies. Strains were taken from mating compatible monokaryons from Michigan collections and fruited tester strains from the United Kingdom and Edmonton, Canada. Growth rates were measured for three replicate plates of modified Brodie medium for each strain. The plates were inoculated by transferring a 0.5 cm diameter plug to the center of each plate and incubation of the plates for 12 days at 25 °C. Every 4 days the linear extension of a randomly chosen radius from the central plug was measured. Growth rate was estimated by linear regression in values of cm day−1.
Data analysis
For all analyses, we assumed the infinite allele model of evolution in which all alleles are equally related by mutation regardless of size. A Perl script was created to identify repeated multi-locus genotypes allowing for missing data, which was used to generate a clone-corrected data set for estimating inbreeding and population structure. The probability that repeated multilocus genotypes were derived from separate sexual events rather than asexual reproduction was estimated using PSex (f), a modified PSex probability that incorporates FIS into account was estimated using Genclone 2.0 (Parks and Werth, 1993; Arnaud-Haond and Belkhir, 2007). In order to estimate PSex (f) for repeated genotypes within each population, we deleted any loci of a population with missing data. Inbreeding coefficients, expected and observed heterozygosity were estimated using Genepop 4.0.10 (Raymond and Rousset, 1995). Deviation from Hardy–Weinberg equilibrium was tested using the exact test for heterozygote deficiency of Raymond and Rousset (1995) as implemented in Genepop.
Population structure was estimated using a hierarchical analysis of molecular variance (AMOVA) implemented in the program Arlequin 3.5.1.3 (Excoffier and Lischer, 2010). This analysis partitions genetic variation into differences within and among groups and estimates Wright’s fixation indices (Weir and Cockerham, 1984). Values reported herein are averages for locus-by-locus global estimates because of the high variance in fixation indices among loci. The C. stercoreus samples were divided into two regions (Louisiana and Michigan) with three sub-populations in each. This sampling structure allows estimation of the correlation of alleles within an individual relative to the total sample (FIT), the correlation of alleles within an individual because of inbreeding within populations (FIS), the correlation of alleles within an population because of reduced gene flow among populations within regions (FSC) and the correlation of alleles within regions because of reduced gene flow among regions (FCT) (Holsinger and Weir, 2009). Significance of each value was tested using 1000 permutations. Clustering of genotypes into hypothetical populations was estimated by Bayesian analysis with the program STRUCTURE 2.3.3 (Pritchard et al., 2000) using the admixture ancestry models with correlated allele frequencies and sampling locations used as priors. The maximum likelihood best estimate of number of populations was estimated following the method of Evanno et al. (2005), as calculated using STRUCTURE HARVESTER v0.6.8 (Earl and vonHoldt, 2012), from five replicate runs for population numbers between n=1 and n=8 with a burn-in of 50 000 generations and sample size of 500 000 generations; the membership matrix was displayed with the help of distruct (Rosenberg, 2004).
Spatial autocorrelation measures the correlation between genetic relatedness and spatial distance among individuals. We estimated autocorrelation using SPAGeDi 1.3 (Hardy and Vekemans, 2002) with genetic relatedness estimated using the Fij kinship coefficient among genotypes described by (Ritland, 1996). Louisiana and Michigan populations were analyzed separately. The number of and maximum extent of distance classes was determined with the goal of maximizing the number of comparisons for each distance class while extending comparisons across meaningful distances.
Results
Microsatellite marker development
Using the draft genome assembly of CS4.14, we designed primers for 45 potential microsatellite markers for analysis of North American C. stercoreus populations. Each primer pair was tested on a diverse set of 5–11 collections that included the global isolates in order to find the most informative and consistent markers. Of these initial markers, 21 did not amplify for the majority of isolates and were discarded. Of the remaining 24 markers, 8 did not provide clear, interpretable results after labeled fragment analysis. Of the markers with clear microsatellite chromatograms, 5 markers were eliminated because of low polymorphism, leaving 11 markers that were used to genotype the sample of 72 individuals. Markers containing tri and hexa nucleotide repeats were found to have the highest allele diversity (Table 1). We evaluated the genotype data using MICRO-CHECKER, and one marker revealed consistent evidence of homozygote excess suggesting a possible null allele in five of the six populations. This marker also had the greatest amount of missing data (10 individuals with the next highest being 6 and an average of 2), potentially because of null allele homozygotes, and thus it was removed from all further analysis. Four other loci showed evidence of homozygote excess within 1–3 populations. As variance in FIS is often observed when sample sizes are low (Castric et al., 2002; Casado-Amezua et al., 2012), and our specific goal is to detect inbreeding, we retained these loci for further analyses.
Population structure
Three populations in Michigan and three in Louisiana were genotyped to investigate the spatial scale of population structure in C. stercoreus. The markers were polymorphic in all populations with an average of 8.7 alleles per locus. First, the data were inspected for repeated multi-locus genotypes, that is, putative clones. From the 72 genotyped samples, we detected 59 distinct multi-locus genotypes (Table 2). In each case, identical multi-locus genotypes were only found within the same sub-population. Some of the repeated genotypes were collected in close proximity, whereas others were collected up to 380 m distant. The probability that the repeated number of multi-locus genotypes (MLGs) were not clones but actually derived from independent mating events (PSex (f)) was estimated for each of 13 different repeated MLGs, and ranged from (PSex (f)=0.0033–1.76 × 10-9). Thus, the identical MLGs are likely to represent the products of clonal reproduction. To make sure all further statistical analysis would not be biased by repetitive MLGs, duplicate MLGs were removed, leaving the sample in the data set with the least amount of missing data for all following analyses.
The data were investigated for underlying population structure using STRUCTURE 2.3.3 with sampling locations as priors. Both the maximum likelihood estimate of the number of populations and the ΔK statistic (Evanno et al., 2005) suggested two clusters, which primarily separated Michigan and Louisiana samples (Figure 1). This spatial structure was further described using a hierarchical multi-locus analysis of molecular variance, allowing the genetic variation to be partitioned into allele correlations at varying spatial structure. Most of the variation is found within individuals (89.5%), with the remaining variation explained by inbreeding and population structure. The fixation indices estimated by hierarchical analysis of molecular variance were: FIT=0.105; FCT=0.033, FSC=0.013 and FIS=0.061. These data show that only 3.3% of the total variation could be explained by differences between regions, that is, Michigan and Louisiana. An even smaller percentage of variation is due to differences between populations within regions, leaving the majority of the remaining correlation due to differences among individuals within populations (≈6%), that is, inbreeding. All fixation indices were significantly different from zero, except FSC (P=0.057) as measured by a permutation test. When each locus was analyzed independently, a high variation in F-statistics among loci were observed (Table 3).
Graphical display of the posterior probabilities of the STRUCTURE analysis. Shown is the run with the highest posterior probability and K=2. Each bar represents the posterior probability of assignment of a single individual to one of two populations. Data were generated using the correlated-allele frequency model with admixture and sampling locations used as priors. Populations are labeled according to Table 1.
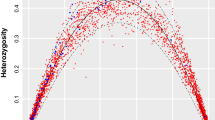
Spatial autocorrelation was used to determine whether there was a relationship between physical distance between collections and genetic distance as predicted under isolation by distance. Coefficients of genetic relatedness were calculated among collections separately for Louisiana and Michigan. The slope of the regression of Fij kinship coefficients on ln (distance) was significantly negative for the Louisiana population (P<0.02; Figure 2a) but not the Michigan population (P>0.5; Figure 2b). These data suggest isolation by distance only for the Louisiana population.
Auto-correlograms of distance by relatedness for Louisiana (a) and Michigan (b) populations. Points represent maximum distance of pairs of samples for that class. Line indicates the best fit line. Error bars indicate s.e.m. Fij values among the 10 loci. The correlation of distance and relatedness was tested by permutation of locations among genotypes, which was significant only for the Louisiana population (P<0.05).
Evidence for inbreeding and inbreeding depression within populations
Evidence of inbreeding was detected as significant heterozygote deficiency within only two sub-populations in Michigan and Louisiana (Table 2). Within Michigan, the Ann Arbor, Lansing and Grand Valley sub-populations all have positive FIS values, although only Ann Arbor showed a significant heterozygote deficiency (P=0.0034). Within Louisiana, all populations had positive FIS values (0.041, 0.105 and 0.006) but only LSU showed significant heterozygote deficiencies (P=0.0206; Table 2).
To investigate whether the detected inbreeding is associated with inbreeding depression, growth rates of a sample of inbred and outbred individuals were measured in the Michigan population. Inbred individuals were derived from selfing spores obtained from the same fruiting body, whereas outbred individuals were constructed from spores isolated from separate fruiting bodies. The observed values of both groups had a nearly identical range of approximately threefold from 0.15–0.45 cm day−1, suggesting complex control of growth rates. The mean linear growth rate of outbred dikaryons (0.36 cm day−1, n=27) was significantly faster (P<0.05, df=43, T-test) than the mean linear growth rate of inbred dikaryons (0.32 cm day−1, n= 18; Figure 3). The magnitude of inbreeding depression (Lande and Schemske, 1985) was estimated to be δ=1−ws/wx=0.156, where ws is the mean fitness of inbred individuals and wx is the mean fitness of outcrossed individuals.
Estimation of number of mating-type alleles
We estimated the number of mating-type alleles in C. stercoreus using over 800 crosses of single-spore isolates from Michigan and one tester strain from Edmonton, Canada. C. stercoreus is tetrapolar with four mating types per collection, and tester strains representing the four mating types were initially obtained by crosses within a fruiting body. Partial incompatibility (shared mating types) was observed between single-basidiospore isolates of 18 different pairs of fruiting bodies (Figure 4). In total, 15 MAT-A alleles were found. Three MAT-A alleles were found twice, and one MAT-A allele was found four times. A total of 13 MAT-B mating-type alleles were recovered. Four MAT-B alleles were present in two fruiting bodies, two MAT-B alleles were present in three fruiting bodies, and the remaining seven alleles were each recovered a single time. Using the maximum likelihood estimate described in O’Donnell and Lawrence (1984), 39.4 MAT-A alleles (95% confidence interval: 21–115) and 24.3 MAT-B alleles (95% confidence interval: 16–52) are predicted.
Results from the interstrain cross of 40 different haploid cultures derived from 12 fruiting bodies collected from Ann Arbor, MI (2,3,12), East Lansing, MI (4,5,6,7,9), Grand Valley, MI (1,8,11) and Edmonton CAN (10).+, compatible mating, A, incompatibility because of common MAT-A allele, B, incompatibility because of common MAT-B allele.
Discussion
In this study, we evaluated the population genetics of C. stercoreus, a bird’s nest fungus whose dispersal strategy packages numerous compatible gametes into a single dispersing peridiole. This strategy predicts an extreme inbreeding potential for a heterothallic fungus. Furthermore, owing to the large size of the peridioles (>1 mm), long distance dispersal was not predicted for the species. The results reported here, however, provide clear evidence that both outcrossing and long distance dispersal are common in the species, as overall levels of population structure and inbreeding are minimal. However, inbreeding was detected at both the level of individuals within populations and in the form of population structure between Michigan and Louisiana. F-statistics estimated using the hierarchical model allowed contributions of allele correlations at various levels to be compared. The predominant form of inbreeding is suggested to be due to mating of related individuals within sub-populations (FIS) and not due to population structure at the regional (FCT) or sub-population level (FSC), because FIS=0.0612>FCT>FSC.
These results add to a growing appreciation of the understudied phenomenon of inbreeding in natural populations of fungi. Why has inbreeding been so poorly detected in fungal populations? A likely explanation is that it simply has not been detected because it has not been investigated (Billiard et al., 2012). Most fungi are haploid and population genetic studies have largely not included dikaryotic or diploid species like C. stercoreus in which inbreeding can be detected as correlation of the alleles within an individual as departure from Hardy–Weinberg equilibrium. On the other hand, evidence for population structure is pervasive throughout the fungi. This occurs despite the common assumption that the light weight spores of fungi have high potential for gene flow among vast geographic spaces. In our study, only ≈3.3% of the genetic correlations could be explained by differences between regions (Louisiana and Michigan) that are separated by over 1000 km, and although there was clear signal of population structure, assignment of all individuals to Louisiana or Michigan origin was not unambiguous (Figure 1).
A general trend that saprotrophic mushrooms have randomly mating populations has been observed (for example, James et al., 1999; Kauserud and Schumacher, 2001; Franzen et al., 2007). Mushroom species represent the largest fruiting body types in fungi and produce a copious number of spores, facilitating long distance dispersal. Yet, the same reproductive forms are produced by ectomycorrhizal and plant pathogenic mushroom species, and here there is clear evidence for inbreeding within populations of multiple species (Gryta et al., 2000; Amend et al., 2010; Baumgartner et al., 2010; Vincenot et al., 2012). Across the fungal kingdom, there is mounting evidence for mixed mating systems with varying degrees of inbreeding and outcrossing (Marra et al., 2004; Knop, 2006; Giraud et al., 2008). Among the important considerations in understanding why inbreeding occurs in these fungal groups is an appreciation for the distribution of suitable habitat, the mean distance between parents and offspring, and the density of individuals. These factors control the probability of both dispersing to a suitable habitat and of finding a mate. Inbreeding in fungi should be favored when either dispersal distances are low relative to substrate availability or when substrates are separated by large distances but the probability of multiple colonization by propagules low. In the latter case, specific adaptations may evolve to generate diploid or dikaryotic propagules that promote diploid selfing. For example, in both Nidulariaceae, the button mushroom Agaricus bisporus, and the anther smut Microbotryum violaceum, the primary dispersal propagule is effectively diploid. The patchy availability of substrates, such as dung, may have selected for codispersal of gametes as a form of reproductive assurance.
Inbreeding depression was detected in C. stercoreus as a significant reduction in the growth rate of inbred dikaryons relative to outcrossed. Inbreeding depression is primarily attributable to the presence of recessive deleterious alleles (Charlesworth and Willis, 2009), and it may occur even in species that frequently undergo inbreeding such as highly selfing flowering plants because of reduction in effective population size (Charlesworth et al., 1990). Although mutations negatively effecting growth rate should be purged from C. stercoreus populations in the haplophase, because the spores of C. stercoreus germinate immediately in the substrate (typically dung) after heat activation, it appears that finding a mate would occur quickly, reducing the duration of the free-living haploid phase and facilitating the sheltering of deleterious alleles. Interestingly, monokaryons of many bird’s nest fungi grow very slowly and haphazardly relative to dikaryons (Brodie, 1948), which may be due to the accumulation of recessive deleterious mutations within populations. The magnitude of inbreeding depression (δ=0.156) is lower than that observed in the majority of studies of wild populations of animals and plants (Crnokrak and Roff, 1999). However, our estimate of inbreeding depression was similar to the same fitness measurement in Agaricus bisporus (δ=0.117) recorded by Xu (1995), a species that also appears to have a mixed mating system that is characterized by codispersal of compatible gametes.
One surprising result of this study was the determination of a much higher number of MAT alleles at both loci in C. stercoreus relative to other previously studied Nidulariaceae (Brodie, 1975). These estimates are still considerably lower than the majority of other investigated mushroom fungi (Raper, 1966). It is possible that evolution of new alleles may be retarded in Nidulariaceae because its mating system and possible absence of a prolonged haplophase have allowed deleterious mutations linked to the mating-type loci to accumulate because of recombination suppression in the MAT locus region (Uyenoyama, 2005). This is supported by data showing biased mating-type segregation from recovered from single-spore isolates in experimental settings (Brodie, 1975). It could also be that the effective population sizes of C. stercoreus have historically been much smaller. C. stercoreus and other Nidulariaceae are dominant in ruderal urban habitats where they thrive in new mulch beds, suggesting their ecological niche has greatly widened in the last two centuries. The urban landscape is high in ultraviolet radiation, high in pollutants, and may generally be considered a patchy and low quality habitat. Two hypotheses may be put forward to explain the prevalence of bird’s nest fungi in mulch beds in urban environments. First, adaptation to the herbivore gut may have selected for traits that are equally beneficial in wood chips. However, not all of the species with a hapteron that allows for attachment to vegetation are associated with herbivore dung, for example, species like C. striatus that are primarily on fallen wood or mulch. This notion, therefore, of herbivore dispersal may be paradigmatic for only a small number of bird’s nest fungi, and perhaps may be only one of several means of dispersal. Second, the process of generating mulch may somehow be selecting or adding C. stercoreus during either harvesting or production. For example, because C. stercoreus germinate following high temperature shock, the species may be among the earliest colonizers of the mulch if it is piled and composted. Given that C. stercoreus has a higher number of mating-type alleles than other Nidulariaceae, it would be of interest to investigate the DNA sequences of the mating-type loci to determine if they have undergone recent allele diversification as the result of its population expansion into urban environments.
The populations from Louisiana differed in a number of ways from those of Michigan. Louisiana had slightly higher diversity than Michigan as measured by HE. A major difference, however, was the higher frequency of clonal genotypes in Louisiana relative to Michigan and the significant spatial autocorrelation that was only observed in Louisiana. This observation creates a paradox as clonality can be best explained by human-mediated dispersal, whereas spatial autocorrelation is typically the result of natural dispersal processes. Many of the clonal genotypes were adjacent, however, at least one repeated MLG in Louisiana occurred in patches separated by 380 m. Non-adjacent clones in Louisiana argues for either a colonization of the mulch before its application to the landscape or for movement of the mycelium by humans or other animals after application.
Despite these differences between Louisiana and Michigan population structures, the genetic differentiation between the two regions is modest (FCT=0.033). This low level of differentiation between the two populations is similar to other mushroom fungi sampled at a similar scale that show remarkably homogenous populations across very different habitats (Kauserud and Schumacher, 2003; Baumgartner et al., 2010; Vincenot et al., 2012). In these cited studies, the implication is that microscopic spores often <20 μm are dispersed as bioaerosols over long distances. For bird’s nest fungi like C. stercoreus, it is unclear how the large peridioles could be dispersed long distances. Possible explanations for this low population differentiation include a recent range expansion of the species through association of the species with urbanization. In this scenario, the populations have yet to reach migration-drift equilibrium. An alternative is that herbivores such as horses may have spread the spores over long distances either presently or in the recent past when horse transport was the dominant mode of travel in North America.
Although the results support a significant level of inbreeding, not all of the loci supported this hypothesis (Table 3). For neutral loci (e.g., microsatellites) in obligately sexual populations, this result is not expected. Variance in FIS among loci could be attributed to stochastic founder effects related to the number of parental genotypes contributing gametes during the colonization of these isolated urban mulched areas (Castric et al., 2002). We suggest that because the loci showing significant departures from Hardy–Weinberg equilibrium in the various sub-populations differed, the variance in FIS among loci is due to founder effects. Unfortunately, owing to the small sample sizes of the populations we were unable to test for founder effects or bottlenecks as deviation in allele frequency distribution (excess of intermediate allele frequencies).
Data archiving
Genotype data have been deposited in the Dryad repository: doi:10.5061/dryad.0h054.
References
Amend A, Garbelotto M, Fang ZD, Keeley S (2010). Isolation by landscape in populations of a prized edible mushroom Tricholoma matsutake. Conserv Genet 11: 795–802.
Arnaud-Haond S, Belkhir K (2007). GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7: 15–17.
Baumgartner K, Travadon R, Bruhn J, Bergemann SE (2010). Contrasting patterns of genetic diversity and population structure of Armillaria mellea sensu stricto in the eastern and western United States. Phytopathology 100: 708–718.
Billiard S, Lopez-Villavicencio M, Hood ME, Giraud T (2012). Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol 25: 1020–1038.
Brodie HJ (1948). Tetrapolarity and unilateral diploidization in the bird's nest fungus Cyathus stercoreus. Am J Bot 35: 312–320.
Brodie HJ (1951). The splash-cup dispersal mechanism in plants. Can J Bot-Rev Can Bot 29: 224–234.
Brodie HJ (1975) The Bird’s Nest Fungi. University of Toronto Press: Toronto.
Casado-Amezua P, Goffredo S, Templado J, Machordom A (2012). Genetic assessment of population structure and connectivity in the threatened Mediterranean coral Astroides calycularis (Scleractinia, Dendrophylliidae) at different spatial scales. Mol Ecol 21: 3671–3685.
Castric V, Bernatchez L, Belkhir K, Bonhomme F (2002). Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus Fontinalis Mitchill (Pisces, Salmonidae): a test of alternative hypotheses. Heredity 89: 27–35.
Charlesworth B, Charlesworth D (1999). The genetic basis of inbreeding depression. Genet Res 74: 329–340.
Charlesworth B, Charlesworth D, Morgan MT (1990). Genetic loads and estimates of mutation rates in highly inbred plant populations. Nature 347: 380–382.
Charlesworth D, Willis JH (2009). The genetics of inbreeding depression. Nat Rev Genet 10: 783–796.
Crnokrak P, Roff DA (1999). Inbreeding depression in the wild. Heredity 83: 260–270.
deVisser J, Hoekstra RF, vandenEnde H (1997). Test of interaction between genetic markers that affect fitness in Aspergillus niger. Evolution 51: 1499–1505.
Earl DA, vonHoldt BM (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4: 359–361.
Eugenio CP, Anderson NA (1968). Genetics and cultivation of Pleurotus ostreatus. Mycologia 60: 627.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Excoffier L, Lischer HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567.
Flegler SL (1979). Improved method for production of Cyathus stercoreus fruit bodies in culture. Mycologia 71: 625–627.
Franzen I, Vasaitis R, Penttila R, Stenlid J (2007). Population genetics of the wood-decay fungus Phlebia centrifuga P Karst in fragmented and continuous habitats. Mol Ecol 16: 3326–3333.
Fries N (1940). Researches into the multipolar sexuality of Cyathus striatus. Symb Bot Upsaliens 4: 1–39.
Fries N (1943). Über das Vorkommen von geographischen Rassen bei Crucibulum vulgare. Archiv für Mikrobiol 13: 182–190.
Giraud T, Yockteng R, Lopez-Villavicencio M, Refregier G, Hood ME (2008). Mating system of the anther smut fungus Microbotryum violaceum: Selfing under heterothallism. Eukaryot Cell 7: 765–775.
Gryta H, Debaud JC, Marmiesse R (2000). Population dynamics of the symbiotic mushroom Hebeloma cylindrosporum: mycelial persistence and inbreeding. Heredity 84: 294–302.
Hardy OJ, Vekemans X (2002). SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Holsinger KE, Weir BS (2009). Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet 10: 639–650.
Ingold CT (1953) Dispersal in Fungi. Oxford University Press: Oxford.
James TY, Porter D, Hamrick JL, Vilgalys R (1999). Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune. Evolution 53: 1665–1677.
James TY, Stenlid J, Olson A, Johannesson H (2008). Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution 62: 2279–2296.
Kauserud H, Schumacher T (2001). Outcrossing or inbreeding: DNA markers provide evidence for type of reproductive mode in Phellinus nigrolimitatus (Basidiomycota). Mycol Res 105: 676–683.
Kauserud H, Schumacher T (2003). Genetic structure of Fennoscandian populations of the threatened wood-decay fungus Fomitopsis rosea (Basidiomycota). Mycol Res 107: 155–163.
Keller LF, Waller DM (2002). Inbreeding effects in wild populations. Trends Ecol Evol 17: 230–241.
Knop M (2006). Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. Bioessays 28: 696–708.
Kües U, James TY, Heitman J (2011). Mating type in Basidiomycetes: unipolar, bipolar, and tetrapolar patterns of sexuality. In: Pöggeler S, Wöstemeyer J (eds) Evolution of Fungi and Fungal-Like Organisms, The Mycota XIV. Springer Verlag: Berlin, pp 97–160.
Lacey J (1996). Spore dispersal—its role in ecology and disease: the British contribution to fungal aerobiology. Mycol Res 100: 641–660.
Lande R, Schemske DW (1985). The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39: 24–40.
Leslie JF, Raju NB (1985). Recessive mutations from natural populations of Neurospora crassa that are expressed in the sexual diplophase. Genetics 111: 759–777.
Marra RE, Cortesi P, Bissegger M, Milgroom MG (2004). Mixed mating in natural populations of the chestnut blight fungus, Cryphonectria parasitica. Heredity 93: 189–195.
O’Donnell S, Lawrence MJ (1984). The population genetics of the self-incompatibility polymorphism in Papaver rhoeas. IV. The estimation of the number of alleles in a population. Heredity 53: 495–507.
Parks JC, Werth CR (1993). A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae). Am J Bot 80: 537–544.
Pringle A, Taylor JW (2002). The fitness of filamentous fungi. Trends Microbiol 10: 474–481.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Raper JR (1966) Genetics of Sexuality in Higher Fungi. Ronald Press: New York.
Raper JR, Raper CA (1968). Genetic regulation of sexual morphogenesis in Schizophyllum commune. J Elisha Mitchell Sci Soc 84: 267–273.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Ritland K (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67: 175–185.
Rosenberg NA (2004). DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138.
Rozen S, Skaletsky HJ (2000). Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press: Totowa, NJ, pp 365–386.
Schoustra SE, Bataillon T, Gifford DR, Kassen R (2009). The properties of adaptive walks in evolving populations of fungus. PLoS Biol 7: e1000250.
Schuelke M (2000). An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18: 233–234.
Simchen G (1967). Genetic control of recombination and the incompatibility system in Schizophyllum commune. Genet Res 9: 195–210.
Thiel T, Michalek W, Varshney RK, Graner A (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106: 411–422.
Uyenoyama MK (2005). Evolution under tight linkage to mating type. New Phytol 165: 63–70.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Vincenot L, Nara K, Sthultz C, Labbe J, Dubois MP, Tedersoo L et al (2012). Extensive gene flow over Europe and possible speciation over Eurasia in the ectomycorrhizal basidiomycete Laccaria amethystina complex. Mol Ecol 21: 281–299.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Wright S (1939). The distribution of self-steriliity alleles in populations. Genetics 24: 538–552.
Xu JP (1995). Analysis of inbreeding depression in Agaricus bisporus. Genetics 141: 137–145.
Brown AJ, Casselton LA (2001). Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet 17: 393–400.
Zerbino DR, Birney E (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829.
Acknowledgements
We thank Marissa Antosiewicz, Courtney Frye and Maria Lee for technical assistance. We also thank Brendan Tarrier at the University of Michigan Sequencing Core for his help in preparing Illumina libraries for sequencing. Cathie Aime and her research group provided generous help in identifying sampling localities in Louisiana. We thank two anonymous reviewers for helpful suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Malloure, B., James, T. Inbreeding depression in urban environments of the bird’s nest fungus Cyathus stercoreus (Nidulariaceae: Basidiomycota). Heredity 110, 355–362 (2013). https://doi.org/10.1038/hdy.2012.95
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/hdy.2012.95
Keywords
This article is cited by
-
Population structure of an orchid mycorrhizal fungus with genus-wide specificity
Scientific Reports (2017)