Abstract
Narrow endemics are at risk from climate change because of their restricted habitat preferences, lower colonization ability and dispersal distances. Landscape genetics combines new tools and analyses that allow us to test how both past and present landscape features have facilitated or hindered previous range expansion and local migration patterns, and thereby identifying potential limitations to future range shifts. We have compared current and historic habitat corridors in Cirsium pitcheri, an endemic of the linear dune ecosystem of the Great Lakes, to determine the relative contributions of contemporary migration and post-glacial range expansion on genetic structure. We used seven microsatellite loci to characterize the genetic structure for 24 populations of Cirsium pitcheri, spanning the center to periphery of the range. We tested genetic distance against different measures of geographic distance and landscape permeability, based on contemporary and historic landscape features. We found moderate genetic structure (Fst=0.14), and a north–south pattern to the distribution of genetic diversity and inbreeding, with northern populations having the highest diversity and lowest levels of inbreeding. High allelic diversity, small average pairwise distances and mixed genetic clusters identified in Structure suggest that populations in the center of the range represent the point of entry to the Lake Michigan and a refugium of diversity for this species. A strong association between genetic distances and lake-level changes suggests that historic lake fluctuations best explain the broad geographic patterns, and sandy habitat best explains local patterns of movement.
Similar content being viewed by others
Introduction
Under current projections of climate change, many plant species are expected to experience a shift in distribution. Narrow endemics, often characterized by limited ranges, restricted habitat preferences and variable population size (Rabinowitz, 1981; Lavergne et al., 2004), have been identified as a group at risk of extinction from climate change (Damschen et al., 2010). The range of narrow endemics is often restricted by the distribution and connectivity of appropriate habitats (Alvarez et al., 2009; Meirmans et al., 2011). In cases where the edges of the distribution are determined by habitat features other than climatic tolerances, a species may still be able to persist in their current location by adaptation to the new climate regime or through plasticity (Nicotra et al., 2010). However, for species in which climate is an important determinant of range limits, climate change will likely result in a contraction of the current range. Alternatively, if climate conditions have restricted a species’ ability to colonize new habitats outside the current range, the rate of expansion will depend on dispersal potential. For these reasons, the conservation of rare narrow endemics will require a better understanding of how their current distributions have been influenced by colonization patterns and previous range shift.
The distribution of genetic diversity throughout a plant’s range is dependent on life-history traits (Hamrick and Godt, 1996; Nybom, 2004), in particular, pollination and dispersal mechanisms (Theil-Egenter et al., 2009; Kramer et al., 2011; Meirmans et al., 2011), available habitat (Alvarez et al., 2009; Meirmans et al., 2011) and historical events (Hewitt, 2000; Petit et al., 2003, 2008; Hu et al., 2009). Glaciers covered the upper Midwest of North America until 14 000 yBP (Colman et al., 1994b); hence, most of the region’s present day vegetation migrated in from unglaciated areas (Gleason, 1922; McLaughlin, 1932). Migration routes used varied with species’ climatic and soil preferences (Gleason, 1922; McLaughlin, 1932; Curtis, 1959), with many local endemic plants, which persist in extreme habitats such as xerophytic sites or bogs, representing relic colonies of post-glacial migration patterns (Gleason, 1922). The species that grow on the dunes and beaches of the Great Lakes are thought to have migrated from the Atlantic coast entering the region from the northeast, along sand deposits associated with post-glacial lakes and drainage channels (McLaughlin, 1932; Reznicek, 1994). Alternatively, these sand endemic species may have migrated from the Southern Coastal plains up the Mississippi River, entering the Great Lakes from the southern tip of Lake Michigan (Gleason, 1922; McLaughlin, 1932; Curtis, 1959). A third possibility is that these species are derived from the unglaciated Nebraska Sandhills to the southwest, although this route would require high dispersal distances given the lack of continuous sandy habitat (Gleason, 1922; McLaughlin, 1932; Shapiro, 1971).
Cirsium pitcheri (Eaton) Torrey and Gray (Pitcher’s thistle or dune thistle) is a federally listed rare species and endemic of the shorelines of the Great Lakes of North America. The highest density of C. pitcheri populations is along the northern shores of Lake Michigan, with populations becoming sparser and more spread out toward the western and southern edges of Lake Michigan and on the Canadian and American shores of Lakes Superior and Huron (Guire and Voss, 1963).The high density of populations to the northeast would suggest that this area represents the rear edge of the species’ entry into the region. However, the likely closest congener of C. pitcheri is C. canescens from the Nebraska Sandhills (Moore and Frankton, 1963), which might support a more southwesterly origin. A previous range-wide study using isozymes suggested that diversity was slightly higher in the northeastern edge of the range but was hindered by insufficient polymorphisms to draw any major conclusions (Loveless and Hamrick, 1988).
Populations at the range edge are at higher risk under most climate change scenarios, consequently distinguishing between expanding edge, and rear edge is important for the conservation of a species (Hampe and Petit, 2005). Recent microsatellite studies focusing on small populations of C. pitcheri at the northern and southern extremes of the range have shown low diversity, high inbreeding and low connectivity between populations (Gauthier et al., 2010; Fant et al., 2013). These results may represent founding edge populations that have just a subset of the diversity from the historic origin of the species (Bialozyt et al., 2006; Hu et al., 2009), alternatively this pattern may be a consequence of these populations having smaller, more widely dispersed and lower densities associated with less suitable habitat (Eckert et al., 2008; Sexton et al., 2009). The persistence of C. pitcheri in an area depends on a cycle of the local extinctions associated with loss of habitat from succession, and colonization of newly opened sandy habitat generated by geological and aeolian processes (Loveless, 1984; McEachern et al., 1994). Given the fragmented nature of C. pitcheri habitat, short lifespan, low dispersal probability and early successional status, the low connectivity seen in the northern and southern edges of the range may have important consequences for future migration potential.
Here, we use the microsatellite markers (Fant et al., 2013) to characterize range-wide patterns in the distribution of genetic diversity within this species. Given the poor dispersal and low connectivity in this species (Fant et al., 2013), we hypothesize that populations that represent the rear edge would have maintained higher diversity compared with populations at the expanding edge. We also hypothesize that the highest diversity will likely be in the northeast of Lake Michigan, which is thought to be the point of entry of many dune specialists from the Atlantic Coast. At more local scale, we hypothesize that connectivity between populations will depend on contemporary landscape features, such as distance between neighboring populations, habitat size and availability, and coastal barriers. New tools and analyses developed for landscape genetic studies now allow researchers to quantify and test the effects of landscape features in driving population genetic structure (McRae and Beier, 2007; Cushman and Landguth, 2010; Holderegger et al., 2010; Storfer et al., 2010). In this study we propose to investigate genetic discontinuities in the context of contemporary and historic landscape features, such as soil type and changes in lake levels. In combination, these factors will be quantified to assess their influence on patterns of gene flow and population genetic structure in this narrow endemic species and identify potential limitations to future range shifts, given climate change.
Materials and methods
Study species
C. pitcheri is a short-lived, monocarpic, herbaceous plant, generally flowering after a 4- to 8-year juvenile stage (Loveless, 1984). Individuals typically have a single, branching flowering stem with terminal and axillary flowering heads of cream or pinkish color. C. pitcheri is insect-pollinated and believed to be partially self-incompatible (Bowles, McEachern and Pavlovic, pers. obs.). It appears to have a very short-lived seed bank (Loveless, 1984; McEachern, 1992; Bowles and McBride, 1996; Hamzé and Jolls, 2000), which is unlikely to contribute significantly to population growth (Rowland and Maun, 2001).
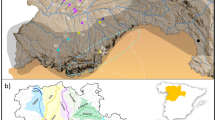
C. pitcheri is found on both the Canadian and American shores of Lakes Superior and Huron (Guire and Voss, 1963), although the majority of the 203 known occurrences are along the shores of Lake Michigan (Figure 1). Of these, at least 18 have been extirpated, including all occurrences in Illinois. The sporadic distribution of C. pitcheri makes it difficult to delineate biological populations. Here, we follow the definition by Pavlovic et al. (2002) of a population, as an element occurrence as groups of individuals separated from other such groups by one mile. Based on this definition, population sizes vary throughout the range from fewer than 100 individuals to more than 10 000 individuals. Given the life history of the species, only a small proportion of individuals will flower within any 1 year, therefore effective population sizes are likely to be considerably smaller than the census size (Vitalis et al., 2004).
The distribution of Cirsium pitcheri. Circles represent all known elemental occurrences, dark circles in Canada represent populations from Gauthier et al. (2010), and stars are populations included in this study.
Study sites
Twenty-four sites were selected that represented the four habitat types (narrow linear foredunes, continuous complex dunes, discontinuous complex dunes and perched dunes) and the five geographic locations defined by Loveless and Hamrick (southern Lake Michigan (SLM), northern Lake Michigan (NLM), Wisconsin, Upper Peninsula Michigan (Lake Superior, LS) and Straits of Mackinac (SM); Table 1; Figure 1). An additional three populations were added to the six populations sampled in Fant et al. (2013) for a total of eight populations from SLM, 10 populations were added from northern Lake Michigan, in addition, two populations were sampled from the SM, two populations from the LS and one population from the Lake Huron (LH) (Table 1) for a total of 24 populations (Table 1). Collections were made over an 8-year period from 2005 to 2013 (Table 1). Size classes, rather than counts, were used to account for seasonal fluctuation. Size classes and the dune-habitat type definitions were derived from Pavlovic et al. (2002). A measurement of the degree of isolation of each population was calculated as the average Euclidean distance to the 5, 10 and 20 nearest populations, using Geographic Distance Matrix Generator (version 1.2.3) (Ersts, 2013). As the three measurements showed no significant difference from each other, we only used the nearest 20 populations for all analyses. Correlations between population attributes (dune type, population size and average distance to nearest populations) and location (longitude and latitude) were calculated using R statistical package (R Development Core Team, 2009).
Molecular data
Leaf tissue collections were made within 100 m of the center of the elemental occurrences to minimize potential Wahlund effect associated with collecting from distant patches (Pavlovic et al., 2002; Fant et al., 2013). In larger populations (>200 plants), leaf samples were collected haphazardly from 50 individuals, although to avoid sampling siblings we did not collect samples from adjacent plants (within a 5-m radius). In smaller populations, leaf samples were collected from every plant with more than five adult leaves. At all study sites, 1–5 g of fresh leaf tissue were collected and dried in silica gel for later DNA extraction.
Total genomic DNA was extracted from silica-dried leaf material using Qiagen DNeasy kits (Qiagen Inc., Valencia, CA, USA). DNA quantity was visually estimated on a gel using standards and diluted to a final concentration of 5 μg ml−1. Seven microsatellite loci were amplified from all individuals using the polymerase chain reaction and fluorescently tagged forward primers (WellRed D2, D3 or D4, Sigma-Proligo, St Louis, MO, USA) described in Fant et al. (2013). An additional primer (Caca23) that was modified from Jump et al., 2002 (Genbank accession AJ457857) was found to be polymorphic across the ranges and was included in this study (forward: 5′-TTGAACCCTTTTGAAGCACA-3′ and reverse: 5′-CACCCAGAAACATAACGGATT-3′). Subsets of individuals from the larger populations (Pictured Rocks, Wilderness State Park and Sleeping Bear Dunes) were re-extracted and genotyped by a second individual to test for repeatability. Dewoody et al. (2006) identified 2% as a reasonable error rate, which is not likely to bias data analysis. The genotyping error was <2% for all markers, except Caca17 that was at a little under 4%. The genotyping error in Caca17 was associated with mis-identification of new alleles; hence, all samples with rare alleles were rerun. All primers were tested for each locus, population and globally for the potential of null alleles and mis-scoring using exact tests in Micro-Checker (van Oosterhout et al., 2004).
Geographic data
To help identify important drivers of the genetic discontinuities within C. pitcheri, four different measures of geographic distance and habitat permeability were calculated to test isolation by distance. Euclidean distance, calculated as the shortest distance between two points using ESRI ArcGIS 10 (ESRI, 2011), represents the simplest model as it assumes that there are no geographic or biological barriers preventing gene flow between any population pairs. Shoreline distance assumes that the lake is a barrier to gene flow; therefore, the least cost path for gene flow is restricted to the thin narrow dune habitat along the coast. To calculate the distance between populations along the shoreline, we used Network Analyst extension of ESRI ArcGIS 10, which creates an Origin-Destination cost matrix using the Great Lakes shoreline boundary (GLIN, 2011) to measure pairwise distances. As a sand specialist, C. pitcheri requires sandy habitat to support its expansion into the area. We used Circuitscape 3.5 (McRae, 2006) and widely available GIS maps to calculate the permeability of the landscape based on sand availability. The open sandy habitat that C. pitcheri requires is created by either shoreline erosion associated with tidal movement, lake-level drops or deposits from glacial retreat and was lost through succession or lake-level rises. Soil maps (Harmonized World Soils Database, 1 km resolution; IIASA-ISRIC-ISS-CAS –JRC, 2009) were used to identify potential sites with sufficient sand that could have historically supported C. pitcheri (Loope and McEachern, 1998; Larson and Schaetzl, 2001). Circuitscape uses electrical circuit theory to determine the path of least resistance between sites by applying weights to landscape variables based on the likelihood of facilitating or preventing movement and connectivity (McRae, 2006; Shah and McRae, 2008).Using the soil raster data set, any soil that had high sand composition (>75%; based on composition of soil at sites of known occurrence) was assigned a low resistance (cell value=1). Other soil types were considered to have too low a sand content to have supported C. pitcheri populations, and therefore were assigned infinite resistance (cell value=no data). Open water was assigned a higher resistance than sandy soil types, to allow for the possibility that it is not a complete barrier to gene flow (cell value=4).
To account for habitat availability associated with historic drops in the lake level, we used bathometry layers (NOAA, National Oceanic and Atmospheric Administration, National Geophysical Data Center (NGDC), 2011). Lake Michigan has seen two dramatic drops in lake levels; the first was a rapid draining of the lake associated with the opening of the SM 10 000 yBP (Schaetzl et al., 2002) and the second was a rapid decline 7000 yBP associated with a period of warmer and drier climate (Colman et al., 1994a, 1994b). The second decline was the lowest level the lake reached, estimated to be a drop of 100 m below current levels (Colman et al., 1994b). Using the bathometry raster, lake-level depths of 0–100 m below the current level were assigned a low resistance to represent possible habitat and corridors opened when lake levels dropped (cell value=1). The remaining open water was assigned a higher resistance (cell value=4) and non sandy land mass was assigned infinite resistance (cell value=no data). Input data were projected to the US Contiguous Lambert Azimuthal Equal Area projection and the Spatial Analyst extension in ESRI ArcGIS 10 was utilized to reclassify raster input layers (ESRI, 2011). The rasters are input into Circuitscape, which calculates pairwise resistances and creates maps of current flowing between focal nodes (that is, study species populations).
Statistical analysis
Genetic variation
Descriptive parameters include the following: average number of samples genotyped across all loci, accounting for missing data (N), mean number of alleles per locus (Ap), expected heterozygosity (He) and the number of monomorphic loci; the proportion of known alleles found in this population (%A) and Weir and Cockerham’s (1984) estimates of Wright’s FIS (within population-inbreeding coefficient) were calculated in GENALEX (Peakall and Smouse, 2006). Allelic richness, adjusted for sample size, was calculated in FSTAT (Goudet, 1995).
Backward elimination of generalized-linear models was used to test for significance of population size, habitat type, isolation (average distance of 20 closest populations), latitude and longitude on each measure of genetic diversity (Ap, He, allelic richness and %A) and Weir and Cockerham’s (1984) estimates of Wright’s FIS. Correlations and generalized-linear model were calculated and tested for significance using R statistical package (R Development Core Team, 2009). BOTTLENECK v1.2.0 was used to test for evidence of recent and past bottlenecks (Cornuet and Luikart, 1997) using a Wilcoxon’s signs test to look for evidence of heterozygous excess and deficiency using both the Infinite Allele Model and the Two-Phased Model of mutation, a variant of the strict Stepwise Mutation Model (Luikart et al., 1998). The Two-Phased Model of mutation was run for 105 simulations with 95% single-step mutations and 5% multi-step mutations and a variance of 12 as recommended by Piry et al. (1999) and Chiucchi and Gibbs (2010). As these first tests are best for detecting bottlenecks in the 0.2–0.4 Ne generations, we also employed the mode-shift test for detecting more recent bottlenecks (Cornuet and Luikart, 1997; Luikart et al., 1998; Chiucchi and Gibbs, 2010).
Population differentiation
The Bayesian clustering analysis software STRUCTURE v2.2 (Pritchard et al., 2000; Falush et al., 2007) was used to visualize patterns of gene flow (admixture, Q) and population subdivision (number of genetic clusters, K) among study populations. This software uses individual multilocus genotypes to test for the presence of population structure without a priori assignment of individuals to populations by introducing population structure and finding population groupings with the least possible disequilibrium (HWE and linkage disequilibrium) using a Markov–Chain Monte Carlo method. We carried out 20 independent runs per K using a burn-in period of 105 and collected data for 105 iterations for K=1–30. The minimum value of K that can explain the data was assessed using the rate of change in the log likelihood probability of data between corresponding K values (ΔK) as detailed in Evanno et al. (2005).
Isolation by distance
SPAGeDi (Hardy and Vekemans, 2002) was used to calculate three measures of pairwise genetic distance among populations: (1) Nei’s (1978) standard genetic distance (Ds) (Hardy et al., 2003, 2) Weir and Cockerham FST (Weir and Cockerham, 1984), and (3) Rousset’s linearized FST (FST /(1−FST); Rousset, 1997), all of which assume that each mutation can produce an allele of any size and hence differences between populations are driven primarily by drift. Although GST (Hedrick, 2005) and a new statistic, D (Jost, 2008) are thought to circumvent some statistical problems with FST, recent studies have affirmed that FST and its equivalents are still better statistical measure for making demographic inferences (Meirmans and Hedrick, 2011; Meirmans et al., 2011; Whitlock, 2011).
To test for isolation by distance, pairwise genetic distances were regressed against the four measures of spatial distance including the log of Euclidean distance (km), log of shoreline distance (km) and two measures of habitat permeability as calculated in Circuitscape. The correlation between genetic structure and the four measures of geographic distance, as well as correlation between each measure of geographic distance, was determined using Mantel (1967) tests (103 permutations) in GENALEX (Peakall and Smouse, 2006). Partial Mantel tests (103 permutations) were conducted in XLSTAT-Pro (Statistical Innovations, MA) (Smouse et al., 1986) to identify spurious correlations from potential explanatory variables, as described in Cushman and Landguth (2010). This was first ran with all population pairs, and then to test for potential migration routes was repeated with only population pairs from south to center of the range (Sleeping bear Dunes) and then again with those pairs from center to north of the range.
Results
Descriptive statistics of populations
There was a negative correlation between latitude and population size (r=−0.45; P=0.02) and longitude and population size (r=−0.55; P=0.008), which supports the observation that populations in the western and southern edges of the range are smaller than those in the northern and eastern edges. Interestingly, degree of isolation was not correlated with population size or latitude but was negatively on correlated with longitude (r=−0.61; P=0.002), likely driven by the scarcity of populations along the Wisconsin coast on the western edge of the range. Population size varied by dune type, with perched dunes, which have large areas of open sand, supporting larger populations, whereas discontinuous dunes with restricted open sand, supporting smaller populations. Continuous and linear dunes ranged from large to mid-range sized populations. Perched dunes are restricted to Picture Rocks National Lakeshore in the North and Sleeping Bear Dunes National Lakeshore in the center of the range, and linear dunes are not common in the southern half of the range, although some of the extinct populations in Illinois were historically on linear dunes. Both continuous and discontinuous dunes were sampled throughout the range. There was no significant relationship between isolation and dune type.
Measurements of genetic diversity varied considerably by population. Average number of alleles per loci (Ap) ranged from 1.3 to 4.0, gene diversity (He) ranged from 0.13 to 0.52, allelic richness ranged from 1.3 to 3.0, and proportion of known alleles (%A) ranged from 31 to 74% (Table 1). Backward elimination of generalized-linear models suggested that dune type, longitude and log of isolation were not good predictors of any of the four measures of genetic diversity. However, population size showed a positive relationship for three of the four measures of diversity (Ap(t1,20=3.6, P=0.002), He (t1,17=2.7, P=0.01) and allelic richness (t1,20=3.3, P=0.003)) (Figure 2a), whereas latitude showed a positive relationship for all measures of genetic diversity (Ap(t1,20=2.8, P=0.02), He (t1,17=3.7, P=0.001), allelic richness (t1,20=3.2, P=0.004) (Figure 2b) and %A (t1,21=4.9, P=0.002)).
(a) Average allelic richness by population size class (<100 plants, 100–500 plants, 500–5000 plants, 5000–10 000 and >10 000 plants), with line of best fit and correlation in parenthesis. (b) Average allelic richness by latitude with line of best fit and correlation in parenthesis. (c) Proportion of known alleles by population size class (<100 plants, 100–500 plants, 500–5000 plants, 5000–10 000 and >10 000 plants) with correlation in parenthesis. (d) Proportion of known alleles by latitude with line of best fit and correlation in parenthesis. (e) Inbreeding coefficient (Fis) by population size class (<100 plants, 100–500 plants, 500–5000 plants, 5000–10 000 and >10 000 plants) with correlation in parenthesis. (f) Inbreeding coefficient (Fis) by latitude with line of best fit and correlation in parenthesis.
Weir and Cockerham’s (1984) estimates of Wright’s FIS ranged from no evidence of inbreeding (−0.03) to relatively high levels of inbreeding (0.33) (Table 1). Backward elimination suggests that the only significant predictor for inbreeding was latitude, which had a significant negative relationship (t1,20=−3.0, P=0.003; Figure 2d). No significant bottleneck was detected using either the Stepwise Mutation Model or Two-Phased Model of mutation models in BOTTLENECK v1.2.0 (Cornuet and Luikart, 1997; Table 1). However, three populations (NLM-WI2, NLM-UP3 and LH-HSP1) did show a mode-shift in distribution, from the L shape expected under mutation-drift equilibrium (data not shown) suggesting evidence of a recent bottleneck (Cornuet and Luikart, 1997; Luikart et al., 1998; Chiucchi and Gibbs, 2010).
Analysis of population genetic structure
Structure 2.2 results confirmed pronounced genetic structure, which differed from that described by Loveless and Hamrick (1988). The modal value of the distribution of the true K identified a peak at ΔK=4, which was supported by large shifts in L(K) and Ln’(K) from K=4 to K=5 associated with true value of K, as described in Evanno et al. (2005). Comparing the proportion of each cluster (K) assigned to populations, Structure identified a geographic gradient, radiating out from Sleeping Bear Dune populations in central Lake Michigan. The individuals from populations in Sleeping Bear Dune National Lakeshore, MI, USA comprised all four cluster types (Figures 3 and 4), with the first cluster becoming more prominent in populations further north. Two of the clusters became increasingly more prominent the further south from Sleeping Bear, with SLM-WIN1 and SLM-WIN3, in western Indiana, being predominately composed of one of the two clusters and the remaining populations in the Eastern half of Indiana and South Michigan being composed of the second. A fourth cluster becomes increasingly more prominent in the Wisconsin populations terminating in NLM-WI3, which is solely identified by this cluster (Figures 3 and 4).
Identified genetic clusters (1–4, as shown at the bottom of the figure) and Bayesian admixture proportions depicted for individual plants and populations spanning the complete range of Cirsium pitcheri. Population names correspond to study site information (Table 1). A full color version of this figure is available at the Heredity journal online.
Isolation by distance
Pairwise genetic distances (FST) ranged from low (0.01) to very high (0.58), with an average of  =0.14 (Table 2). The average pairwise distances by population ranged from 0.09 to 0.33. Most populations were between 0.12 and 0.20, with the exceptions of populations at Sleeping Bear Dunes and on LH, which had the lowest averages (
=0.14 (Table 2). The average pairwise distances by population ranged from 0.09 to 0.33. Most populations were between 0.12 and 0.20, with the exceptions of populations at Sleeping Bear Dunes and on LH, which had the lowest averages ( =0.09–0.10), and NLM-WI3 in Wisconsin that had the highest (
=0.09–0.10), and NLM-WI3 in Wisconsin that had the highest ( =0.33). Mantel (1967) tests of pairwise genetic distances (FST) showed a significant positive correlation to all measures of geographic distances, including log of Euclidian distance (r=0.38, P<0.0001), log of shoreline distance (r=0.58, P<0.0001), and Circuitscape resistance for soil (r=0.51, P<0.0001) and the lake level (r=0.70 P<0.0001) (Figure 5).
=0.33). Mantel (1967) tests of pairwise genetic distances (FST) showed a significant positive correlation to all measures of geographic distances, including log of Euclidian distance (r=0.38, P<0.0001), log of shoreline distance (r=0.58, P<0.0001), and Circuitscape resistance for soil (r=0.51, P<0.0001) and the lake level (r=0.70 P<0.0001) (Figure 5).
Isolation by distance (IBD) comparing pairwise genetic distances (FST) against four measures of geographic distance, Euclidean distance (a), shoreline distance (b) and resistance to movement, calculated in Circuitscape (first using just terrestrial sandy habitat (c) and then historical lake levels (d)). Pairwise distances involving populations at range edge are represented by circles (Western Indianan populations=open circles and Eastern Indiana populations=closed circles) and the remaining pairwise comparisons are represented as diamonds. Map shows (a) Range map showing proximity of populations, (b) Lake perimeter, (c) and (d) show resistance as calculated by Circuitscape grading from red, indicating low resistance to movement, to blue indicating high resistance. A full color version of this figure is available at the Heredity journal online.
As there were significant correlations between the four measures of geographic distance and landscape permeability (ranging from r=0.58 to 0.71; P<0.0001), a partial Mantel test was used to identify spurious from causal correlation, by comparing genetic distance to each geographic distances and landscape permeability measures, while partialling out other potentially covarying variables (Cushman and Landguth, 2010) (Table 3). Euclidean distance was nonsignificant or negative when other measures were partialled out. Shoreline distance and soil layer remained significantly positive, except when the lake-level resistance layer was partialled out. Lake-level change was the only factor that remained significant regardless of which other factor was partialled out, suggesting that the resistance associated with lake-level changes was the best explanatory variable for the genetic distances in these populations. A similar pattern was found when comparisons were restricted to central and northern populations (excluding Indiana and Southern Michigan population), suggesting that lake-level changes were the best explanation of genetic distance between the north and center of the range. When the comparison was repeated for populations from central and southern populations, Euclidean and shoreline distance and lake-level resistance were not significantly correlated with genetic distance. There was a significant positive correlation between soil layer and genetic distance (r=0.24, P<0.0001), which suggests that available inland sandy habitat is the best explanatory variable for migration from the center of the range to the south.
Local population pairs (that is, separated by distances <1–50 km; Table 2) showed low to moderate pairwise genetic distance (FST=0.01–0.05) in the central populations (Sleeping Bear Dunes) but moderate genetic distance between southern population pairs (Indiana populations) (FST=0.04–0.18) and between northern population pairs (LS, Upper Peninsula and Straits of Mackinaw) (0.06–0.12). Comparison of genetic distances and all measures of geographic distances produced a significant but weak positive correlation in central populations for the log of Euclidian distance (r=0.30, P<0.0001), log of shoreline distance (r=0.31, P<0.0001) and Circuitscape resistance for the lake level (r=0.20, P<0.0001) but large significant correlation for resistance by soil type (r=0.73, P<0.0001). None of the correlations were significant for southern or northern populations.
Discussion
Genetic structure within C. pitcheri revealed a geographic divide, with populations in the extreme northern, western, southeastern and southwestern edges of the range being distinct. These genetic groupings differ from those described by Loveless and Hamrick (1988) with SM, Upper Peninsula, LH and LS forming a single genetic cluster in our study. Populations in Sleeping Bear Dunes, at the center of the range, were identified in Structure as being comprised of a combination of all four genetic clusters and they showed the smallest average pairwise distances to all other populations suggesting that they are intermediate to all other populations. There was also a strong geographic pattern to the distribution of genetic diversity, with latitude being correlated with all measures of diversity including proportion of all known alleles, suggesting that northern and central populations hold more of the known allelic diversity. The low diversity in the south and in the Canadian populations, at the far northern end of the range (Gauthier et al., 2010), supports the center–periphery model, with diversity being highest in the center of the range and declining toward the edges (Brown, 1984; Hampe and Petit, 2005). The populations at the northern and southern range edges are smaller and at lower densities; however, we found no evidence of greater isolation to the south than those at the core of the species range (Pavlovic et al., 2002). As genetic diversity was also correlated with population size, a pattern found in other endemic Cirsium species (Gauthier et al., 2010; Jacquemyn et al., 2010), the geographic pattern to genetic diversity could be driven by smaller populations along the edges of the range. However, as the proportion of known alleles within a population was not correlated with the size but was with the latitude, this gives support to a geographic pattern to diversity. Inbreeding was also correlated to latitude but not to population size or degree or isolation; with populations at the southern edge still showed higher inbreeding. This is surprising as we found no evidence that populations were more isolated at range edges, yet the pairwise genetic distances in Indiana are some of the highest seen range wide.
C. pitcheri migrated into its current range post glaciation. A survey of Lake Michigan has shown that C. pitcheri populations currently occupy, or have occupied, much of the available habitat in the area (Pavlovic et al., 2002); hence, the likely drivers of range limits are the presence of suitable habitat and successional competition rather than dispersal limitations (McEachern, 1992; Sexton et al., 2009). The two most likely entry points for this species are from the northeast, the site of glacial Lake Arkona, along the Atlantic Coastal route or from the south, the site of glacial Lake Chicago, along the Southern Coastal plains along the Mississippi (Gleason, 1922; McLaughlin, 1932; Curtis, 1959; Larson and Schaetzl, 2001). The high allelic diversity at the northeastern edge of the range and the low diversity at the southern edge suggests that the northeast represents the refugia of diversity for this species and the most likely point of entry to the region (Petit et al., 2003; Hampe and Petit, 2005). This is further supported by the Structure analysis that reveals multiple genetic clusters within central populations and small average pairwise genetic distances of the central region to all other populations. This is not surprising, given that Michigan’s Lower Peninsula was one of the first areas to be exposed after glacial melt; its high sand content and access to multiple lakes make it an ideal migration route and refuge for C. pitcheri during the large fluctuations in the water levels of the Great Lakes (Larson and Schaetzl, 2001).
Lake-level changes were the best explanation of genetic structure in C. pitcheri, especially for population in the north and center of the range, rather than contemporary shoreline, Euclidean distance or sandy-soil availability. This suggests that previous lake-level drops represent a time of greater connectivity especially in the northern edge of range between C. pitcheri populations either through pollen or seed movement, or range expansion because of the availability of new sandy habitat. The largest drop in the lake level occurred at 7000 yBP, and the lake did not return to current levels for another 2000 years (Colman et al., 1994b).The lake level peaked around 4500 yBP at 10 m higher than current levels, which would have reduced available habitat and increased isolation of the surviving populations (Colman et al., 1994b; Thompson et al., 2011).
Genetic distance from the southern edge of range to central populations was not explained by either Euclidean distance, shoreline distance or changes in Lake level; however, there was weak association with available sandy substrate. This might suggest that populations along the southern edge of the range might have been larger or had greater connectivity in the past, although much of this habitat has been lost to C. pitcheri through succession (Cowles, 1899). Sandy habitat was also the best explanatory variable for genetic distance in the central region at a local scale (<50 km). This is not surprising as we know that population dynamics of C. pitcheri is driven by a cycle of the local extinctions, associated with loss of habitat from succession and colonization of new open sandy habitat generated by geological and aeolian processes (Loveless, 1984; McEachern et al., 1994). The lack of correlation between Euclidean and shoreline distance, moderate pairwise genetic distances and high inbreeding in the populations in SLM suggests poor conductivity and high degree of isolation in these populations, despite relatively small distance. A similar pattern was found in 20-year-old restorations where populations <500 m apart showed no evidence of introgression (Fant et al., 2013). Although this high divergence may be exacerbated by recent (>150 years) increases in fragmentation because of development in the area, the magnitude of difference is greater than can be expected, given the size of populations and generation time (Lloyd et al., 2013), suggesting that the divergence is in part an historic legacy.
The relatively small genetic distance between populations at the southern edge of range to those in Sleeping Bear Dunes National Lakeshore, and given the southern populations only contain a subset of known alleles, would suggest that these populations are the result of sporadic founding events from the range center, rather than the result of a genetic bottleneck at the retreating edge (Hewitt, 2000; Petit et al., 2003, 2008; Hu et al., 2009). The presence of two genetic groups in Indiana might suggest separate founding populations, which is not surprising given the dunes in the western portion of Indiana are younger, less stable and wider than those in the east of Indiana (Olson, 1958; Cowles, 1899). A similar pattern of multiple independent origins was observed in local haplotypes of Panicum virgatum in the same region (Morris et al., 2011). By contrast, Kohler–Andrae State Park (NLM-WI3) at the western edge of the range, which is the most isolated C. pitcheri population sampled, had high levels of inbreeding and low diversity but showed evidence of a recent genetic bottleneck, suggesting that this population might be a result of the retreating range edge or product of a limited number of founders.
C. pitcheri with lower colonization ability and dispersal distances, similar to many narrow endemics (Fiedler, 1987; Byers and Meagher, 1997; Lloyd et al., 2003), shows a spatial genetic structure resulting from a combination of contemporary isolation associated with habitat availability at a local scale and rare sporadic long-distance migration events associated with range expansion. The strong association between genetic distances and historic lake-level changes suggests that periods of lake fluctuations might best explain the broad geographic patterns. High genetic diversity of C. pitcheri in the northeast, and the relatively high number of rare dune endemics found in this area, including Houghton’s goldenrod (Solidago houghtonii A. Gray), Dwarf lake iris (Iris lacustris Nutt.) and Lake Huron Tansy (Tanacetum huronense Nutt.), suggests that this region represents the likely original site of entry for C. pitcheri as well as other dune endemics (Guire and Voss, 1963; Hannan and Orick, 2000; Hamilton and Eckert, 2007). The mosaic pattern of genetic diversity would indicate that colonization has been sporadic, particularly toward the south, with rare and erratic long-distance dispersal events (Cruzan and Templeton, 2000; Clark, 1998) associated with habitat preference having an important role (Alvarez et al., 2009). For these peripheral populations, the small population sizes, contemporary isolation, demographic decline and poor recruitment may interact to increase the loss of genetic diversity (Schaal and Leverich, 1996), with the potential to induce genetic bottlenecks in the future (Lesica and Allendorf, 1995). Future climate models predict that the southern and western edges of the range will no longer have a suitable climate to support C. pitcheri (Vitt et al., 2010), suggesting a contraction of the distribution of this species back to the historic center of distribution, increasing the conservation value of the dune habitat along the northeast coast of Lake Michigan.
Data Archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.cf165.
Accession codes
References
Alvarez N, Theil-Egenter C, Tribsch A, Holderegger R, Manel S, Schönswetter P et al. (2009). History or ecology? Substrate type as a major driver of spatial genetic structure in alpine plants. Ecol Lett 12: 632–640.
Bialozyt R, Ziegenhagen B, Petit RJ . (2006). Contrasting effects of long distance seed dispersal on genetic diversity during range expansion. J Evol Biol 19: 12–20.
Bowles ML, McBride JL . (1996). Pitcher’s thistle (Cirsium pitcheri) reintroduction. In Falk DA, Millar CI, Olwell M, (eds). Restoring Diversity: Strategies for Reintroduction of Endangered Plants. Island Press: Washington, D.C. pp 423–431.
Brown JH . (1984). On the relationship between abundance and distribution of species. Am Nat 124: 255–279.
Byers DL, Meagher TR . (1997). A comparison of demographic characteristics in a rare and common species of Eupatorium. Ecol Appl 7: 519–530.
Chiucchi JE, Gibbs HL . (2010). Similarity of contemporary and historical gene flow among highly fragmented populations of an endangered rattlesnake. Mol Ecol 19: 5345–5358.
Clark JS . (1998). Why trees migrate so fast: confronting theory with dispersal biology and the paleorecord. Am Nat 152: 204–224.
Colman SM, Keigwinand LD, Forester RM . (1994a). Two episodes of meltwater influx from glacial Lake Agassiz into the Lake Michigan basin: contrasts in climatic and oceanographic effects. Geology 22: 547–550.
Colman SM, Forester RM, Reynolds RL, Sweetkind DS, King JW, Gangemi P et al. (1994b). Lake-level history of Lake Michigan for the past 12000 Years: the record from deep lacustrine sediments. J Great Lakes Res 20: 73–92.
Cornuet JM, Luikart G . (1997). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014.
Cowles HC . (1899). The ecological relations of vegetation on the sand dunes of Lake Michigan. Bot Gazette 27: 95–117.
Cruzan MB, Templeton AR . (2000). Paleoecology and coalescence: phylogeographic analysis of hypotheses from the fossil record. Trends Ecol Environ 15: 491–496.
Curtis JT . (1959) The Vegetation of Wisconsin. University of Wisconsin Press: Madison, WI, USA.
Cushman SA, Landguth EL . (2010). Spurious correlations and inference in landscape genetics. Mol Ecol 19: 3592–3602.
Damschen EI, Harrison S, Grace JB . (2010). Climate change effects on an endemic-rich edaphic flora: resurveying Robert H Whittaker's Siskiyou sites (Oregon, USA). Ecology 91: 3609–3619.
Dewoody J, Nason JD, Hipkins VD . (2006). Mitigating scoring errors in microsatellite data from wild populations. Mol Ecol Notes 6: 951–957.
Eckert CG, Samis KE, Lougheed SC . (2008). Genetic variation across species’ geographical ranges: the central–marginal hypothesis and beyond. Mol Ecol 17: 1170–1188.
Ersts PJ . (2013). Geographic distance matrix generator (version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation. http://biodiversityinformatics.amnh.org/open_source/gdmg.
ESRI. (2011) ArcGIS Desktop: Release 10. Environmental Systems Research Institute: Redlands, CA, USA.
Evanno G, Regnaut S, Gouder J . (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14: 2611–2620.
Falush D, Stephens M, Pritchard JK . (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578.
Fant JB, Kramer AT, Sirkin E, Havens K . (2013). Genetics of reintroduced populations of the narrowly endemic thistle, Cirsium pitcheri (Asteraceae). Botany (online early).
IIASA -ISRIC -ISS-CAS -JRC. (2009) Harmonized World Soil Database (version 1.1). FAO: Rome, Italy and IIASA, Laxenburg, Austria.
Fiedler P. L. . (1987). Life history and population dynamics of rare and common mariposa lilies (Calochortus pursh: Liliaceae). J Ecol 75: 977–995.
Gauthier M, Crowe E, Hawke L, Emery N, Wilson P, Freeland J . (2010). Conservation genetics of Pitcher's thistle (Cirsium pitcheri), an endangered Great Lakes endemic. Botany 88: 250–257.
Gleason HA . (1922). The vegetation history of the middle west. Ann Assoc Am Geogr 12: 29–85.
GLIN. (2011) Great Lakes Information Network. NOAA shoreline layer, 70 km http://gis.glin.net/.
Goudet J . (1995). FSTAT Version 1.2: a computer program to calculate F-statistics. J Heredity 86: 485–486.
Guire KE, Voss EG . (1963). Distributions of distinctive shoreline plants in the Great Lakes region. Mich Bot 2: 99–114.
Hamilton JA, Eckert CG . (2007). Population genetic consequences of geographic disjunction a prairie plant isolated on Great Lakes alvars. Mol Ecol 16: 1649–1660.
Hampe A, Petit RJ . (2005). Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8: 461–467.
Hamrick JL, Godt MJW . (1996). Effects of life history traits on genetic diversity in plant species. Philos T R Soc B 351: 1291–1298.
Hamzé SI, Jolls CL . (2000). Germination ecology of a federally threatened endemic thistle of the Great Lakes, Cirsium pitcheri. Am Midl Nat 143: 141–153.
Hannan GL, Orick MW . (2000). Isozyme diversity in Iris cristata and the threatened glacial endemic I. lacustris (Iridaceae). Am J Bot 8: 293–301.
Hardy OJ, Vekemans X . (2002). SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Hardy OJ, Charbonnel N, Fréville H, Heuertz M . (2003). Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163: 1467–1482.
Hedrick PW . (2005). A standardized genetic differentiation measure. Evolution 59: 1633–1638.
Hewitt GM . (2000). The genetic legacy of the Quaternary ice ages. Nature 405: 907–913.
Holderegger R, Buehler D, Gugerli F, Manel S . (2010). Landscape genetics of plants. Trends Plant Sci 15: 675–683.
Hu FS, Hampe A, Petit RJ . (2009). Paleoecology meets genetics: deciphering past vegetational dynamics. Front Ecol Environ 7: 371–379.
Jacquemyn H, Roldán-Ruiz I, Honnay O . (2010). Evidence for demographic bottlenecks and limited gene flow leading to low genetic diversity in a rare thistle. Conserv Genet 11: 1979–1987.
Jost L . (2008). GST and its relatives do not measure differentiation. Mol Ecol 17: 4015–4026.
Jump AS, Dawson DA, James CM, Woodward FI, Burke T . (2002). Isolation of polymorphic microsatellites in the stemless thistle (Cirsium acaule) and their utility in other Cirsium species. Mol Ecol Notes 2: 589–592.
Kramer AT, Fant JB, Ashley M . (2011). Influences of landscape and pollinators on population genetic structure: examples from three Penstemon (Plantaginaceae) species in the Great Basin. Am J Bot 98: 109–121.
Larson G, Schaetzl RJ . (2001). Origin and evolution of the Great Lakes. J Great Lakes Res 27: 518–546.
Lavergne S, Thompson JD, Garnier E, Debussche M . (2004). The biology and ecology of narrow endemic and widespread plants: a comparative study of trait variation in 20 congeneric pairs. Oikos 107: 505–518.
Lesica P, Allendorf FW . (1995). When are peripheral populations valuable for conservation? Conserv Biol 9: 753–760.
Lloyd KM, Wilson JB, Lee WG . (2003). Correlates of geographic range size in New Zealand Chionochloa (Poaceae) species. J Biogeogr 30: 1751–1761.
Lloyd MW, Campbell L, Neel MC . (2013). The power to detect recent fragmentation events using genetic differentiation methods. PLoS One 8: e63981.
Loope WL, McEachern K . (1998). Habitat change in a perched dune system along Lake Superior. In: Mac MJ, Opler PA, Puckett Haecker CE, Doran PD, (eds). Status and Trends of the Nation's Biological Resources. Biological Resources Division, U.S. Geological Survey: Reston, Virginia.
Loveless MD . (1984) Population Biology and Genetic Organization in Cirsium pitcheri, an Endemic Thistle. Ph.D. Dissertation, University of Kansas: Lawrence.
Loveless MD, Hamrick JL . (1988). Genetic organization and evolutionary history in two North American species of Cirsium. Evolution 42: 254–265.
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB . (1998). Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered 89: 238–247.
Mantel N . (1967). The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220.
McEachern AK . (1992) Disturbance Dynamics of Pitcher’s Thistle (Cirsium pitcheri) Populations in Great Lakes Sand Dune Landscapes. Ph.D. Dissertation, University of Wisconsin-Madison: Madison, WI, USA.
McEachern AK, Bowles ML, Pavlovic NB . (1994). A metapopulation approach to Pitcher’s thistle (Cirsium pitcheri) recovery in southern Lake Michigan dunes. In: Bowles M L, Whelan CJ, (eds). Restoration of Endangered Species: Conceptual Issues, Planning, and Implementation. Cambridge University Press: Cambridge, UK. pp 194–218.
McLaughlin WT . (1932). Atlantic coastal plain plants in the sand barrens of northwestern Wisconsin. Ecol Monogr 2: 335–383.
McRae BH . (2006). Isolation by resistance. Evolution 60: 1551–1561.
McRae BH, Beier P . (2007). Circuit theory predicts gene flow in plant and animal populations. PNAS 104: 19885–19890.
Meirmans PG, Goudet J IntraBioDiv Consortium Gaggiotti O. E. . (2011). Ecology and life history affect different aspects of the population structure of 27 high-alpine plants. Mol Ecol 29: 3144–3155.
Meirmans PG, Hedrick P . (2011). Assessing population structure: FST and related measures. Mol Ecol Res 11: 5–8.
Moore RJ, Frankton C . (1963). Cytotaxonomic notes on some Cirsium species of the Western United States. Can J Bot 41: 1553–1567.
Morris GP, Grabowski P, Borevitz JO . (2011). Genomic diversity in switchgrass (Panicum virgatum): from the continental scale to a dune landscape. Mol Ecol 23: 4938–4952.
Nei M . (1978). Estimation of average heterozygosity and genetic distance for small number of individuals. Genetics 89: 583–590.
Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci 15: 684–692.
NOAA, National Oceanic and Atmospheric Administration, National Geophysical Data Center (NGDC). (2011) Great Lakes Bathymetry, Accessed.
Nybom H . (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13: 1143–1155.
Olson JS . (1958). Rates of succession and soil changes on Southern Lake Michigan. Bot Gaz 119: 125–170.
Pavlovic NB,, Bowles ML, Crispin S, Gibson T, Herman K, Kavetsky R et al. (1993) Pitcher's Thistle (Cirsium pitcheri) Recovery Plan. U.S. Department of the Interior, Fish and Wildlife Service: Minneapolis, MN, USA.
Pavlovic NB, Bowles ML, Crispin S, Gibson T, Herman K, Kavetsky R et al. (2002) Pitchers's Thistle (Cirsium pitcheri) Recovery Plan. U.S. Department of the Interior, Fish and Wildlife Service: Minneapolis, MN, USA.
Peakall R, Smouse PE . (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R et al. (2003). Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300: 1563–1565.
Petit RJ, Hu FS, Dick CW . (2008). Forests of the past; a window to future changes. Science 230: 1450–1452.
Piry S, Luikart G, Cornuet JM . (1999). BOTTLENCK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90: 502–503.
Pritchard JK, Stephens M, Donnely P . (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945.
R Development Core Team. (2009). R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria.
Rabinowitz D . (1981). Seven forms of rarity. In: Synge H, (ed). The Biological Aspects of Rare Plant Conservation. Wiley: New York. pp 205–217.
Reznicek AA . (1994). The disjuct coastal plain flora in the Great Lakes Region. Biol Conserv 68: 203–215.
Rousset F . (1997). Genetic differentiation and estimation of gene flow from F-Statistics under isolation by distance. Genetics 145: 1219–1228.
Rowland J, Maun MA . (2001). Restoration ecology of an endangered plant species: establishment of new populations of Cirsium pitcheri. Restor Ecol 9: 60–70.
Schaal BA, Leverich WJ . (1996). Molecular variation in isolated plant populations. Plant Species Biol 11: 33–40.
Schaetzl RJ, Drzyzga SA, Weisenborn BN, Kincare KA, Lepczyk XC, Shein KA et al. (2002). Measurement, correlation, and mapping of glacial Lake Algonquin shorelines in northern Michigan. Ann Assoc Am Geogr 92: 399–415.
Sexton JP, McIntyre PJ, Angert AL, Rice KJ . (2009). Evolution and ecology of species range limits. Annu Rev Ecol Evol Syst 40: 415–436.
Shah VB, McRae B . (2008). Circuitscape: a tool for landscape ecology. In: Varoquaux G, Vaught T, Millman J, (eds) Proceedings of the 7th Python in Science Conference. SciPy 2008, pp 62–66.
Shapiro AM . (1971). Postglacial geography and the distribution of Poanes viator (Hesperiidae) and other Marsh butterflies. J Res Lepidoptera 9: 125–155.
Smouse PE, Long JC, Sokal RR . (1986). Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst Zool 35: 627–632.
Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP . (2010). Landscape genetics: where are we now? Mol Ecol 19: 3496–3514.
Theil-Egenter C, Gugerli F, Alvarez N, Brodbeck S, Cieslak E, Colli L et al. (2009). Effects of species traits on the genetic diversity of high-mountain plants: a multi-species study across the Alps and the Carpathians. Global Ecol Biogeogr 18: 78–87.
Thompson TA, Lepper K, Endres AL, Johnston JW, Baedke SJ, Argyilan EP et al. (2011). Mid-Holocene lake level and shoreline behavior during the Nipissing phase of the upper Great Lakes at Alpena, Michigan, USA. J Great Lakes Res 37: 567–576.
van Oosterhout C, Hutchinson W, Wills D, Shipley P . (2004). Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–553.
Vitalis R, Glémin S, Olivieri I . (2004). When genes go to sleep: The population genetic consequences of seed dormancy and monocarpic perenniality. Am Nat 163: 295–311.
Vitt P, Havens K, Kramer AT, Sollenberger D, Yates E . (2010). Assisted migration of plants: changes in latitudes, changes in attitude. Biol Conserv 143: 18–27.
Weir BS, Cockerham CC . (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Whitlock MC . (2011). G’ST and D do not replace FST. Mol Ecol 20: 1083–1091.
Acknowledgements
We thank C Flower, K Hartley, L Jefferson, C Jolls, K Ksiazek, B Kramer, J McLain, M Pennacchio, J Phillips, N Sokal and E Sirkin for field and lab assistance. We also wish to thank D Larkin and S Still for their help with the analyses and M Ashley, A Kramer, K Skogen, Skogen Lab and two anonymous reviewers for helpful comments on this manuscript. This research was supported by NSF- LTREB 0516058 to K Havens.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fant, J., Havens, K., Keller, J. et al. The influence of contemporary and historic landscape features on the genetic structure of the sand dune endemic, Cirsium pitcheri (Asteraceae). Heredity 112, 519–530 (2014). https://doi.org/10.1038/hdy.2013.134
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/hdy.2013.134
Keywords
This article is cited by
-
Genetic diversity and population structure of Robinia pseudoacacia from six improved variety bases in China as revealed by simple sequence repeat markers
Journal of Forestry Research (2022)
-
High genomic diversity maintained by populations of Carex scirpoidea subsp. convoluta, a paraphyletic Great Lakes ecotype
Conservation Genetics (2021)
-
Genetic differentiation and diversity of two sympatric subspecies of Castilleja affinis; a comparison between the endangered serpentine endemic (spp. neglecta) and its widespread congener (ssp. affinis)
Conservation Genetics (2018)
-
Persistence of the gypsophile Lepidospartum burgessii (Asteraceae) through clonal growth and limited gene flow
Conservation Genetics (2016)