Abstract
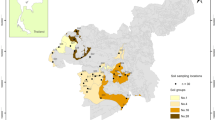
Estimates of the number of species of bacteria per gram of soil vary between 2000 and 8.3 million (Gans et al., 2005; Schloss and Handelsman, 2006). The highest estimate suggests that the number may be so large as to be impractical to test by amplification and sequencing of the highly conserved 16S rRNA gene from soil DNA (Gans et al., 2005). Here we present the use of high throughput DNA pyrosequencing and statistical inference to assess bacterial diversity in four soils across a large transect of the western hemisphere. The number of bacterial 16S rRNA sequences obtained from each site varied from 26 140 to 53 533. The most abundant bacterial groups in all four soils were the Bacteroidetes, Betaproteobacteria and Alphaproteobacteria. Using three estimators of diversity, the maximum number of unique sequences (operational taxonomic units roughly corresponding to the species level) never exceeded 52 000 in these soils at the lowest level of dissimilarity. Furthermore, the bacterial diversity of the forest soil was phylum rich compared to the agricultural soils, which are species rich but phylum poor. The forest site also showed far less diversity of the Archaea with only 0.009% of all sequences from that site being from this group as opposed to 4%–12% of the sequences from the three agricultural sites. This work is the most comprehensive examination to date of bacterial diversity in soil and suggests that agricultural management of soil may significantly influence the diversity of bacteria and archaea.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Aref S, Wander MM . (1998). Long-term trends of corn yield and soil organic matter in different crop sequences and soil fertility treatments on the Morrow Plots. Adv Agron 62: 153–202.
Baker GC, Smith JJ, Cowan DA . (2003). Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55: 541–555.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Keller K, Huber T et al. (2006). Greengenes, a chimera-checked 16S rRNA database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Felsenstein J . (1989). PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166.
Felsenstein J . (2005). PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington: Seattle.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci (USA) 103: 626–631.
Gans J, Woilinsky M, Dunbar J . (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309: 1387–1390.
Hong S-H, Bunge J, Jeon S-O, Epstein SS . (2006). Predicting microbial species richness. Proc Natl Acad Sci USA 103: 117–122.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
Moreira D . (1998). Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res 26: 3309–3310.
Neufeld JD, Mohn WW . (2005). Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl Environ Microbiol 71: 5710–5718.
Phillips N . (1985). The Everglades experience: sixty years of progress. EREC Research Report EV-1985-5. Belle Glade.
Rupert D, Cressie N, Carroll RJ . (1989). A transformation/weighting model for estimating Michaelis–Menton parameters. Biometrics 45: 637–656.
Schloss PD, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Schloss PD, Handelsman J . (2006). Toward a census of bacteria in soil. PLOS Computational Biology 2: 786–793.
Stackebrandt E, Goebel BM . (1994). Taxonomic note: a lace for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Internat J Syst Bacteriol 44: 846–849.
Torsvik V, Goksoyr J, Daae FL . (1990). High diversity in DNA of soil bacteria. Appl Environ Microbiol 56: 782–787.
Treusch AH et al. (2005). Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7: 1985–1995.
Acknowledgements
EWT acknowledges support from the Florida Agricultural Experiment Station, the National Science Foundation (MCB-0454030) and the United States Department of Agriculture (National Research Initiative Competitive Grant 2005-35319-16300). GC was supported by the National Science Foundation (DEB-0540745). RRF received support from National Science and Engineering Research Council Discovery Grant. LFWR and FAOC were supported by the Brazilian Ministry of Education (CAPES) for a scholarship concession, the Estate Research Foundation (FAPERGS) and the Brazilian National Research Council (CNPq). We thank the people of the North Caribou Lake First Nation for guidance and permission to sample soil in their traditional territory in Ontario, Canada. We also thank Jennifer Drew for suggestions on the manuscript before submission.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej).
Rights and permissions
About this article
Cite this article
Roesch, L., Fulthorpe, R., Riva, A. et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1, 283–290 (2007). https://doi.org/10.1038/ismej.2007.53
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2007.53
Keywords
This article is cited by
-
Exploring soil bacterial diversity in different micro-vegetational habitats of Dachigam National Park in North-western Himalaya
Scientific Reports (2023)
-
Bacterial type VI secretion system (T6SS): an evolved molecular weapon with diverse functionality
Biotechnology Letters (2023)
-
A Matter of Metals: Copper but Not Cadmium Affects the Microbial Alpha-Diversity of Soils and Sediments — a Meta-analysis
Microbial Ecology (2023)
-
Uncovering the Secrets of Slow-Growing Bacteria in Tropical Savanna Soil Through Isolation and Genomic Analysis
Microbial Ecology (2023)
-
Sorptive and Desorptive Response of Divalent Heavy Metal Ions from EICP-Treated Plastic Fines
Indian Geotechnical Journal (2023)