Abstract
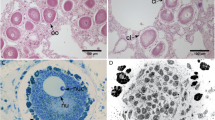
The phylogenetic diversity of microorganisms in marine sponges is becoming increasingly well described, yet relatively little is known about the activities of these symbionts. Given the seemingly favourable environment provided to microbes by their sponge hosts, as indicated by the extraordinarily high abundance of sponge symbionts, we hypothesized that the majority of sponge-associated bacteria are active in situ. To test this hypothesis we compared, for the first time in sponges, 16S rRNA gene- vs 16S rRNA-derived bacterial community profiles to gain insights into symbiont composition and activity, respectively. Clone libraries revealed a highly diverse bacterial community in Ancorina alata, and a much lower diversity in Polymastia sp., which were identified by electron microscopy as a high- and a low-microbial abundance sponge, respectively. Substantial overlap between DNA and RNA libraries was evident at both phylum and phylotype levels, indicating in situ activity for a large fraction of sponge-associated bacteria. This active fraction included uncultivated, sponge-specific lineages within, for example, Actinobacteria, Chloroflexi and Gemmatimonadetes. This study shows the potential of RNA vs DNA comparisons based on the 16S rRNA gene to provide insights into the activity of sponge-associated microorganisms.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Aislabie J, Jordan S, Ayton J, Klassen JL, Barker GM, Turner S . (2009). Bacterial diversity associated with ornithogenic soil of the Ross Sea region, Antarctica. Can J Microbiol 55: 21–36.
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ . (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736.
Bayer K, Schmitt S, Hentschel U . (2008). Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol 10: 2942–2955.
Brinkmann N, Martens R, Tebbe CC . (2008). Origin and diversity of metabolically active gut bacteria from laboratory-bred larvae of Manduca sexta (Sphingidae, Lepidoptera, Insecta). Appl Environ Microbiol 74: 7189–7196.
DeLong EF, Wickham GS, Pace NR . (1989). Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243: 1360–1363.
Diaz MC, Ward BB . (1997). Sponge-mediated nitrification in tropical benthic communities. Mar Ecol Prog Ser 156: 97–107.
Fieseler L, Horn M, Wagner M, Hentschel U . (2004). Discovery of the novel candidate phylum ‘Poribacteria’ in marine sponges. Appl Environ Microbiol 70: 3724–3732.
Friedrich AB, Fischer I, Proksch P, Hacker J, Hentschel U . (2001). Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol Ecol 38: 105–113.
Gentile G, Giuliano L, D′Auria G, Smedile F, Azzaro M, De Domenico M et al. (2006). Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ Microbiol 8: 2150–2161.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95.
Hentschel U, Usher KM, Taylor MW . (2006). Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55: 167–177.
Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J et al. (2002). Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68: 4431–4440.
Hoffmann F, Larsen O, Thiel V, Rapp HT, Pape T, Michaelis W et al. (2005). An anaerobic world in sponges. Geomicrobiol J 22: 1–10.
Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT et al. (2009). Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11: 2228–2243.
Hoffmann F, Roy H, Bayer K, Hentschel U, Pfannkuchen M, Brummer F et al. (2008). Oxygen dynamics and transport in the Mediterranean sponge Aplysina aerophoba. Mar Biol 153: 1257–1264.
Holmes B, Blanch H . (2006). Genus-specific associations of marine sponges with group I crenarchaeotes. Mar Biol 150: 759–772.
Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML . (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4: e1000255.
Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP et al. (1998). Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira like bacteria as dominant populations. Appl Environ Microbiol 64: 3042–3051.
Kane MD, Poulsen LK, Stahl DA . (1993). Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol 59: 682–686.
Longford SR, Tujula NA, Crocetti GR, Holmes AJ, Holmström C, Kjelleberg S et al. (2007). Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat Microb Ecol 48: 217–229.
Ludwig W, Schleifer K-H . (1999). Phylogeny of Bacteria beyond the 16S rRNA standard. ASM News 65: 752–757.
Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J et al. (1998). Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19: 554–568.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Martinez RJ, Mills HJ, Story S, Sobecky PA . (2006). Prokaryotic diversity and metabolically active microbial populations in sediments from an active mud volcano in the Gulf of Mexico. Environ Microbiol 8: 1783–1796.
McIlroy S, Porter K, Seviour RJ, Tillett D . (2008). Simple and safe method for simultaneous isolation of microbial RNA and DNA from problematic populations. Appl Environ Microbiol 74: 6806–6807.
Mills HJ, Martinez RJ, Story S, Sobecky PA . (2005). Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol 71: 3235–3247.
Moeseneder MM, Arrieta JM, Herndl GJ . (2005). A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol Ecol 51: 341–352.
Moeseneder MM, Winter C, Herndl GJ . (2001). Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol Oceanogr 46: 95–107.
Mohamed NM, Colman AS, Tal Y, Hill RT . (2008a). Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ Microbiol 10: 2910–2921.
Mohamed NM, Rao V, Hamann MT, Kelly M, Hill RT . (2008b). Monitoring bacterial diversity of the marine sponge Ircinia strobilina upon transfer into aquaculture. Appl Environ Microbiol 74: 4133–4143.
Mohamed NM, Saito K, Tal Y, Hill RT . (2009). Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J, doi:10.1038/ismej.2009.84.
Morgenroth E, Obermayer A, Arnold E, Brühl A, Wagner M, Wilderer PA . (2000). Effect of long-term idle periods on the performance of sequencing batch reactors. Water Sci Technol 41: 105–113.
Poulsen LK, Ballard G, Stahl DA . (1993). Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol 59: 1354–1360.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Reiswig HM . (1981). Partial carbon and energy budgets of the bacteriosponge Verongia fistularis (Porifera: Demospongiae) in Barbados. PSZNI Mar Ecol 2: 273–293.
Rodriguez-Blanco A, Ghiglione J-F, Catala P, Casamayor EO, Lebaron P . (2009). Spatial comparison of total vs active bacterial populations by coupling genetic fingerprinting and clone library analyses in the NW Mediterranean Sea. FEMS Microbiol Ecol 67: 30–42.
Schloss PD, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U . (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74: 7694–7708.
Schmitt S, Weisz JB, Lindquist N, Hentschel U . (2007). Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl Environ Microbiol 73: 2067–2078.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, de Nys R, Wagner M et al. (2008). Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ Microbiol 10: 1087–1094.
Taylor MW, Hill RT, Piel J, Thacker RW, Hentschel U . (2007a). Soaking it up: the complex lives of marine sponges and their microbial associates. ISME J 1: 187–190.
Taylor MW, Radax R, Steger D, Wagner M . (2007b). Sponge-associated microorganisms: evolution, ecology and biotechnological potential. Microbiol Mol Biol Rev 71: 295–347.
Taylor MW, Schupp PJ, Dahllof I, Kjelleberg S, Steinberg PD . (2004). Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol 6: 121–130.
Thiel V, Neulinger SC, Staufenberger T, Schmaljohann R, Imhoff JF . (2007). Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol Ecol 59: 47–63.
Tringe SG, Hugenholtz P . (2008). A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 11: 442–446.
Troussellier M, Schafer H, Batailler N, Bernard L, Courties C, Lebaron P et al. (2002). Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquat Microb Ecol 28: 13–24.
Vacelet J . (1975). Etude en microscopie electronique de l’association entre bacteries et spongiaires du genre Verongia (Dictyoceratida). J Microsc Biol Cell 23: 271–288.
Vacelet J, Donadey C . (1977). Electron microscope study of the association between some sponges and bacteria. J Exp Mar Biol Ecol 30: 301–314.
Wagner M, Horn M . (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17: 241–249.
Webster NS, Negri AP, Munro MM, Battershill CN . (2004). Diverse microbial communities inhabit Antarctic sponges. Environ Microbiol 6: 288–300.
Webster NS, Wilson KJ, Blackall LL, Hill RT . (2001). Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl Environ Microbiol 67: 434–444.
Weisz JB, Lindquist N, Martens CS . (2008). Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155: 367–376.
West NJ, Obernosterer I, Zemb O, Lebaron P . (2008). Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ Microbiol 10: 738–756.
Wilkinson CR . (1983). Net primary productivity in coral reef sponges. Science 219: 410–412.
Wilkinson CR, Fay P . (1979). Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature 279: 527–529.
Wilkinson CR, Summons RE, Evans E . (1999). Nitrogen fixation in symbiotic marine sponges: ecological significance and difficulties in detection. Mem Queensland Mus 44: 667–673.
Winter C, Moeseneder MM, Herndl GJ . (2001). Impact of UV radiation on bacterioplankton community composition. Appl Environ Microbiol 67: 665–672.
Zhu P, Li Q, Wang G . (2008). Unique microbial signatures of the alien Hawaiian marine sponge Suberites zeteki. Microb Ecol 55: 406–414.
Acknowledgements
We gratefully acknowledge the help of K Lau, G Lear and S Boycheva with RNA analyses, A Turner with TEM, M Mawdsley for assistance with sample collection, and P Deines (all University of Auckland) for helpful discussions. This research was supported by a University of Auckland New Staff Research Fund grant (Project: 9341 3609286) to MWT and a German Research Foundation (DFG) grant (SCHM 2559/1-1) to SS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary information
Rights and permissions
About this article
Cite this article
Kamke, J., Taylor, M. & Schmitt, S. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J 4, 498–508 (2010). https://doi.org/10.1038/ismej.2009.143
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.143
Keywords
This article is cited by
-
Functional conservation of microbial communities determines composition predictability in anaerobic digestion
The ISME Journal (2023)
-
New Negombata species discovered: latrunculin mystery solved
Coral Reefs (2023)
-
Recent Advances of Marine Sponge-Associated Microorganisms as a Source of Commercially Viable Natural Products
Marine Biotechnology (2022)
-
Archaeal communities of low and high microbial abundance sponges inhabiting the remote western Indian Ocean island of Mayotte
Antonie van Leeuwenhoek (2021)
-
Prokaryote Communities Inhabiting Endemic and Newly Discovered Sponges and Octocorals from the Red Sea
Microbial Ecology (2020)