Abstract
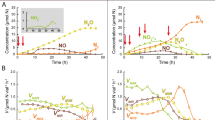
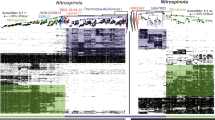
The conversion of nitrite to nitric oxide in the denitrification pathway is catalyzed by at least two structurally dissimilar nitrite reductases, NirS and NirK. Although they are functionally equivalent, a genome with genes encoding both reductases has yet to be found. This exclusivity raises questions about the ecological equivalency of denitrifiers with either nirS or nirK, and how different ecological and evolutionary factors influence community assembly of nirS and nirK denitrifiers. Using phylogeny-based methods for analyzing community structure, we analyzed nirS and nirK data sets compiled from sequence repositories. Global patterns of phylogenetic community structure were determined using Unifrac, whereas community assembly processes were inferred using different community relatedness metrics. Similarities between globally distributed communities for both genes corresponded to similarities in habitat salinity. The majority of communities for both genes were phylogenetically clustered; however, nirK marine communities were more phylogenetically overdispersed than nirK soil communities or nirS communities. A more in-depth analysis was performed using three case studies in which a comparison of nirS and nirK community relatedness within the sites could be examined along environmental gradients. From these studies we observed that nirS communities respond differently to environmental gradients than nirK communities. Although it is difficult to attribute nonrandom patterns of phylogenetic diversity to specific niche-based or neutral assembly processes, our results indicate that coexisting nirS and nirK denitrifier communities are not under the same community assembly rules in different environments.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Abascal F, Zardoya R, Posada D . (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105.
Anonymous . (1958). The Venice system for the classification of marine waters according to salinity. Limnol Oceanogr 3: 346–347.
Braker G, Fesefeldt A, Witzel K . (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64: 3769–3775.
Braker G, Zhou JZ, Wu LY, Devol A, Tiedje JM . (2000). Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities. Appl Environ Microbiol 66: 2096–2104.
Cavender-Bares J, Keen A, Miles B . (2006). Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology 87: S109–S122.
Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW . (2009). The merging of community ecology and phylogenetic biology. Ecol Lett 12: 693–715.
Eddy SR . (1998). Profile hidden Markov models. Bioinformatics 14: 755–763.
Ellis M, Grossman JG, Eady RR, Hasnain SS . (2007). Genomic analysis reveals widespread occurrence of new classes of copper nitrite reductases. J Biol Inorg Chem 12: 1119–1127.
Enwall K, Philippot L, Hallin S . (2005). Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl Environ Microbiol 71: 8335–8343.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci 103: 626–631.
Glockner AB, Jungst A, Zumft WG . (1993). Copper-containing nitrite reductase from Pseudomonas aureofaciens is functional in a mutationally cytochrome-cd1-free background (nirS-) of Pseudomonas stutzeri. Arch Microbiol 160: 18–26.
Gower JC, Legendre P . (1986). Metric and Euclidian properties of dissimilarity coefficients. J Classif 3: 5–48.
Graham CH, Fine PVA . (2008). Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol Lett 11: 1265–1277.
Hallin S, Jones CM, Schloter M, Philippot L . (2009). Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISMEJ 1: 1–9.
Hallin S, Lindgren PE . (1999). PCR detection of genes encoding nitrile reductase in denitrifying bacteria. Appl Environ Microbiol 65: 1652–1657.
Hallin S, Throback IN, Dicksved J, Pell M . (2006). Metabolic profiles and genetic diversity of denitrifying communities in activated sludge after addition of methanol or ethanol. Appl Environ Microbiol 72: 5445–5452.
Horner-Devine MC, Bohannan BJM . (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: S100–S108.
Jones CM, Stres B, Rosenquist M, Hallin S . (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol Bio Evol 25: 1955–1966.
Katoh K, Kuma K, Toh H, Miyata T . (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518.
Kembel SW . (2009). Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol Lett 12: 949–960.
Legendre P, Legendre L . (1998). Numerical Ecology. Elsevier: Amsterdam, The Netherlands.
Letunic I, Bork P . (2007). Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128.
Lozupone CA, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Lozupone CA, Hamady M, Scott KT, Knight R . (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73: 1576–1585.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci 104: 11436–11440.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Martin AP, Costello EK, Meyer AF, Nemergut DR, Schmidt SK . (2004). The rate and pattern of cladogenesis in microbes. Evolution 58: 946–955.
Oakley BB, Francis C, Roberts KJ, Fuchsman CA, Srinivasan S, Staley JT . (2007). Analysis of nitrite reductase (nirK and nirS) genes and cultivation reveal depauperate community of denitrifying bacteria in the Black Sea suboxic zone. Environ Microbiol 9: 118–130.
Paradis E, Claude J, Strimmer K . (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Posada D . (2008). jModelTest: phylogenetic model averaging. Mol Bio Evol 25: 1253–1256.
Pybus O, Rambaut A, Holmes E, Harvey PH . (2002). New inferences from tree shape: numbers of missing taxa and population growth rates. Sys Bio 51: 881–888.
Pybus OG, Harvey PH . (2000). Testing macro-evolutionary models using incomplete molecular phylogenies. Proc R Soc London B Biol Sci 267: 2267–2272.
Rodriguez F, Oliver JL, Marin A, Medina JR . (1990). The general stochastic-model of nucleotide substitution. J Theor Bio 142: 485–501.
Sanderson MJ . (1997). A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Bio Evol 14: 1218–1231.
Santoro AE, Boehm AB, Francis CA . (2006). Denitrifier community composition along a nitrate and salinity gradient in a coastal aquifer. Appl Environ Microbiol 72: 2102–2109.
Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M . (2007). CAMERA: a community resource for metagenomics. PLoS Biol 5: e75.
Smith J, Ogram A . (2008). Genetic and functional variation in denitrifier populations along a short-term restoration chronosequence. Appl Environ Microbiol 74: 5615–5620.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Swofford DL . (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates: Sunderland, MA.
Tuomainen JM, Hietanen S, Kuparinen J, Martikainen PJ, Servomaa K . (2003). Baltic Sea cyanobacterial bloom contains denitrification and nitrification genes, but has negligible denitrification activity. FEMS Microbiol Ecol 45: 83–96.
Webb CO . (2000). Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat 156: 145–155.
Webb CO, Ackerly DD, Kembel SW . (2008). Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24: 2098–2100.
Whelan S, Goldman N . (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Bio Evol 18: 691–699.
Zumft WG . (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Bio Rev 61: 533–616.
Acknowledgements
We thank C Lozupone and S Kembel for their help with software and statistical advice, as well as B Oakley for the additional information on the Black Sea study. We also thank L Philippot for helpful discussions. All phylogenetic analyses were performed using the UPPMAX computational center (Uppsala University, Sweden). Funding by the Swedish Research Council and the Uppsala Microbiomics Center grant from Formas is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Jones, C., Hallin, S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J 4, 633–641 (2010). https://doi.org/10.1038/ismej.2009.152
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.152
Keywords
This article is cited by
-
Soil denitrification response to increased urea concentration constrains nitrous oxide emissions in a simulated cattle urine patch
Plant and Soil (2023)
-
Temporal characteristics of algae-denitrifying bacteria co-occurrence patterns and denitrifier assembly in epiphytic biofilms on submerged macrophytes in Caohai Lake, SW China
Journal of Oceanology and Limnology (2023)
-
Reactive nitrogen restructures and weakens microbial controls of soil N2O emissions
Communications Biology (2022)
-
Impacts of graphitic nanofertilizers on nitrogen cycling in a sandy, agricultural soil
Journal of Nanoparticle Research (2022)
-
Land use in urban areas impacts the composition of soil bacterial communities involved in nitrogen cycling. A case study from Lefkosia (Nicosia) Cyprus
Scientific Reports (2021)