Abstract
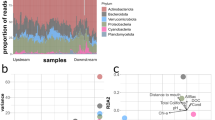
Laboratory studies have documented the extensive architectural differentiation of biofilms into complex structures, including filamentous streamers generated by turbulent flow. Still, it remains elusive whether this spatial organization of natural biofilms is reflected in the community structure. We analyzed bacterial community differentiation between the base and streamers (filamentous structures floating in the water) of stream biofilms under various flow conditions using denaturing gradient gel electrophoresis (DGGE) and sequencing. Fourth-corner analysis showed pronounced deviation from random community structure suggesting that streamers constitute a more competitive zone within the biofilm than its base. The same analysis also showed members of the α-Proteobacteria and Gemmatimonadetes to preferentially colonize the biofilm base, whereas β-Proteobacteria and Bacteroidetes were comparatively strong competitors in the streamers. We suggest this micro-scale differentiation as a response to the environmental dynamics in natural ecosystems.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Besemer K, Singer G, Limberger R, Chlup AK, Hochedlinger G, Hödl I et al. (2007). Biophysical controls on community succession in stream biofilms. Appl Environ Microbiol 73: 4966–4974.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM . (1995). Microbial biofilms. Annu Rev Microbiol 49: 711–745.
Dray S, Legendre P . (2008). Testing the species traits-environment relationships: the fourth-corner problem revisited. Ecology 89: 3400–3412.
Hall-Stoodley L, Costerton JW, Stoodley P . (2004). Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108.
Hallberg KB, Coupland K, Kimura S, Johnson DB . (2006). Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol 72: 2022–2030.
Jackson CR, Churchill PF, Roden EE . (2001). Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82: 555–566.
Kindaichi T, Ito T, Okabe S . (2004). Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol 70: 1641–1650.
Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T . (2003). Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50: 61–68.
Koh KS, Lam KW, Alhede M, Queck SY, Labbate M, Kjelleberg S et al. (2007). Phenotypic diversification and adaptation of Serratia marcescens MG1 biofilm-derived morphotypes. J Bacteriol 189: 119–130.
Legendre P, Galzin R, Harmelin-Vivien ML . (1997). Relating behavior to habitat: solutions to the fourth-corner problem. Ecology 78: 547–562.
Manz W, Wendt-Potthoff K, Neu TR, Szewzyk U, Lawrence JR . (1999). Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridization and confocal laser scanning microscopy. Microb Ecol 37: 225–237.
Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S . (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci USA 102: 16819–16824.
Nikitin DI, Strömpl C, Oranskaya MS, Abraham W-R . (2004). Phylogeny of the ring-forming bacterium Arcicella aquatica gen. nov., sp. nov. (ex Nikitin et al. 1994), from a freshwater neuston biofilm. Int J Syst Evol Microbiol 54: 681–684.
Pang CM, Liu W-T . (2006). Biological filtration limits carbon availability and affects downstream biofilm formation and community structure. Appl Environ Microbiol 72: 5702–5712.
Rickard AH, Leach SA, Hall LS, Buswell CM, High NJ, Handley PS . (2002). Phylogenetic relationships and coaggregation ability of freshwater biofilm bacteria. Appl Environ Microbiol 68: 3644–3650.
Schauer M, Hahn MW . (2005). Diversity and phylogenetic affiliations of morphologically conspicuous large filamentous bacteria occurring in the pelagic zones of a broad spectrum of freshwater habitats. Appl Environ Microbiol 71: 1931–1940.
Singer G, Besemer K, Hödl I, Chlup AK, Hochedlinger G, Stadler P et al. (2006). Microcosm design and evaluation to study stream microbial biofilms. Limnol Oceanogr Meth 4: 436–447.
Stoodley P, Lewandowski Z, Boyle JD, Lappin-Scott HM . (1999). Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: An in situ investigation of biofilm rheology. Biotechnol Bioeng 65: 83–92.
Venugopalan VP, Kuehn M, Hausner M, Springael D, Wilderer PA, Wuertz S . (2005). Architecture of a nascent Sphingomonas sp. biofilm under varied hydrodynamic conditions. Appl Environ Microbiol 71: 2677–2686.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol 73: 5261–5267.
Whitman WB, Coleman DC, Wiebe WJ . (1998). Prokaryotes: The unseen majority. Proc Natl Acad Sci USA 95: 6578–6583.
Wilmes P, Remis JP, Hwang M, Auer M, Thelen MP, Banfield JF . (2009). Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J 3: 266–270.
Zhang K, Choi H, Dionysiou DD, Sorial GA, Oerther DB . (2006). Identifying pioneer bacterial species responsible for biofouling membrane bioreactors. Environ Microbiol 8: 433–440.
Acknowledgements
We thank G Hochedlinger, A-K Chlup, G Steniczka, C Preiler, C Baranyi, M Roura-Carol, E Sollböck and H Hofreiter for their help in the laboratory and in the field. This research was supported by grants of the Austrian Science Fund (P16935-B03) and the European Science Foundation (COMIX, AI0004321) to TJB.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Besemer, K., Hödl, I., Singer, G. et al. Architectural differentiation reflects bacterial community structure in stream biofilms. ISME J 3, 1318–1324 (2009). https://doi.org/10.1038/ismej.2009.73
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.73
Keywords
This article is cited by
-
Unraveling the biophysical underpinnings to the success of multispecies biofilms in porous environments
The ISME Journal (2019)
-
The ecology and biogeochemistry of stream biofilms
Nature Reviews Microbiology (2016)
-
A survey of biofilms on wastewater aeration diffusers suggests bacterial community composition and function vary by substrate type and time
Applied Microbiology and Biotechnology (2016)
-
A multi-disciplinarily designed mesocosm to address the complex flow-sediment-ecology tripartite relationship on the microscale
Environmental Sciences Europe (2015)
-
Adaptation of intertidal biofilm communities is driven by metal ion and oxidative stresses
Scientific Reports (2013)