Abstract
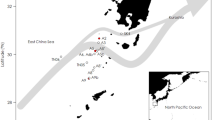
Little is known about the archaeal diversity of fermented seafood; most of the earlier studies of fermented food have focused on lactic acid bacteria (LAB) in the fermentation process. In this study, the archaeal and bacterial diversity in seven kinds of fermented seafood were culture-independently examined using barcoded pyrosequencing and PCR–denaturing gradient gel electrophoresis (DGGE) methods. The multiplex barcoded pyrosequencing was performed in a single run, with multiple samples tagged uniquely by multiplex identifiers, using different primers for Archaea or Bacteria. Because PCR–DGGE analysis is a conventional molecular ecological approach, this analysis was also performed on the same samples and the results were compared with the results of the barcoded pyrosequencing analysis. A total of 13 372 sequences were retrieved from 15 898 pyrosequencing reads and were analyzed to evaluate the diversity of the archaeal and bacterial populations in seafood. The most predominant types of archaea and bacteria identified in the samples included extremely halophilic archaea related to the family Halobacteriaceae; various uncultured mesophilic Crenarchaeota, including Crenarchaeota Group I.1 (CG I.1a and CG I.1b), Marine Benthic Group B (MBG-B), and Miscellaneous Crenarchaeotic Group (MCG); and LAB affiliated with genus Lactobacillus and Weissella. Interestingly, numerous uncultured mesophilic Crenarchaeota groups were as ubiquitous in the fermented seafood as in terrestrial and aquatic niches; the existence of these Crenarchaeota groups has not been reported in any fermented food. These results indicate that the archaeal populations in the fermented seafood analyzed are diverse and include the halophilic and mesophilic groups, and that barcoded pyrosequencing is a promising and cost-effective method for analyzing microbial diversity compared with conventional approaches.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Aller JY, Kemp PF . (2008). Are Archaea inherently less diverse than Bacteria in the same environments? FEMS Microbiol Ecol 65: 74–87.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Ampe F, Ben Omar N, Moizan C, Wacher C, Guyot JP . (1999). Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl Environ Microbiol 65: 5464–5473.
Ampe F, Miambi E . (2000). Cluster analysis, richness and biodiversity indexes derived from denaturing gradient gel electrophoresis fingerprints of bacterial communities demonstrate that traditional maize fermentations are driven by the transformation process. Int J Food Microbiol 60: 91–97.
Ampe F, Sirvent A, Zakhia N . (2001). Dynamics of the microbial community responsible for traditional sour cassava starch fermentation studied by denaturing gradient gel electrophoresis and quantitative rRNA hybridization. Int J Food Microbiol 65: 45–54.
Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L . (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3: e2836.
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ . (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736.
Bae JW, Rhee SK, Park JR, Chung WH, Nam YD, Lee I et al (2005). Development and evaluation of genome-probing microarrays for monitoring lactic acid bacteria. Appl Environ Microbiol 71: 8825–8835.
ben Omar N, Ampe F . (2000). Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl Environ Microbiol 66: 3664–3673.
Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R et al (2007). The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2: e197.
Caplice E, Fitzgerald GF . (1999). Food fermentations: role of microorganisms in food production and preservation. Int J Food Microbiol 50: 131–149.
Casamayor EO, Massana R, Benlloch S, Ovreas L, Diez B, Goddard VJ et al (2002). Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol 4: 338–348.
Cha YJ, Lee EH . (1985). Studies on the processing of low salt fermented sea foods. Bull Korean Fish Soc 18: 206–213.
Chang HW, Kim KH, Nam YD, Roh SW, Kim MS, Jeon CO et al (2008). Analysis of yeast and archaeal population dynamics in kimchi using denaturing gradient gel electrophoresis. Int J Food Microbiol 126: 159–166.
Cheigh HS, Park KY . (1994). Biochemical, microbiological, and nutritional aspects of kimchi (Korean fermented vegetable products). Crit Rev Food Sci Nutr 34: 175–203.
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK et al (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57: 2259–2261.
Cocolin L, Manzano M, Cantoni C, Comi G . (2001). Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl Environ Microbiol 67: 5113–5121.
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM et al (2007). The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172.
Crisan EV, Sands A . (1975). Microflora of four fermented fish sauces. Appl Microbiol 29: 106–108.
DeLong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
DeLong EF, Taylor LT, Marsh TL, Preston CM . (1999). Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol 65: 5554–5563.
DeSantis Jr TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al (2006). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399.
Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA et al (2008a). Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8: 43.
Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA . (2008b). Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis 5: 459–472.
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM et al (2006). Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7: 57.
Ercolini D . (2004). PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Methods 56: 297–314.
Euzeby JP . (1997). List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47: 590–592.
Ferris MJ, Muyzer G, Ward DM . (1996). Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62: 340–346.
Fleet GH . (1999). Microorganisms in food ecosystems. Int J Food Microbiol 50: 101–117.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102: 14683–14688.
Giraffa G . (2004). Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol Rev 28: 251–260.
Giraffa G, Neviani E . (2001). DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int J Food Microbiol 67: 19–34.
Good IJ . (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264.
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J et al (2006a). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA 103: 18296–18301.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al (2006b). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95.
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R . (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237.
Henckel T, Friedrich M, Conrad R . (1999). Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol 65: 1980–1990.
Herrmann M, Saunders AM, Schramm A . (2008). Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl Environ Microbiol 74: 3279–3283.
Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P et al (2007). DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res 35: e91.
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huys G, Vanhoutte T, Joossens M, Mahious AS, De Brandt E, Vermeire S et al (2008). Coamplification of eukaryotic DNA with 16S rRNA gene-based PCR primers: possible consequences for population fingerprinting of complex microbial communities. Curr Microbiol 56: 553–557.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean margin. Proc Natl Acad Sci USA 103: 2815–2820.
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA et al (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57.
Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC et al (2008). Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87: 1016–1020.
Kim BS, Kim BK, Lee JH, Kim M, Lim YW, Chun J . (2008). Rapid phylogenetic dissection of prokaryotic community structure in tidal flat using pyrosequencing. J Microbiol 46: 357–363.
Kumar S, Tamura K, Nei M . (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163.
Lafarge V, Ogier JC, Girard V, Maladen V, Leveau JY, Gruss A et al (2004). Raw cow milk bacterial population shifts attributable to refrigeration. Appl Environ Microbiol 70: 5644–5650.
Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ et al (2007). Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA 104: 7104–7109.
Lee CH . (1994). Importance of lactic acid bacteria in non-dairy food fermentation. In: Lee CH, Nisse JA, Barwald G (eds). Lactic Acid Fermentation of Non-Dairy Food and Beverages. Harnrimwon Publishing CoL: Seoul, Korea. pp 17.
Lee CH . (1997). Lactic acid fermented foods and their benefits in Asia. Food Control 8: 259–269.
Lee CH, Steinkraus KH, Reilly PJA . (1993). Fish Fermentation Technology. United Nations University Press: Tokyo.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Liu Z, DeSantis TZ, Andersen GL, Knight R . (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36: e120.
Lozupone C, Hamady M, Knight R . (2006). UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF . (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9: 1162–1175.
Muyzer G, de Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700.
Nam YD, Chang HW, Kim KH, Roh SW, Kim MS, Jung MJ et al (2008). Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J Microbiol 46: 491–501.
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C . (2003). Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5: 787–797.
Ovreas L, Forney L, Daae FL, Torsvik V . (1997). Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63: 3367–3373.
Paludan-Muller C, Huss HH, Gram L . (1999). Characterization of lactic acid bacteria isolated from a Thai low-salt fermented fish product and the role of garlic as substrate for fermentation. Int J Food Microbiol 46: 219–229.
Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M et al (2007). A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35: e130.
Randazzo CL, Torriani S, Akkermans AD, de Vos WM, Vaughan EE . (2002). Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl Environ Microbiol 68: 1882–1892.
Rivas R, Velazquez E, Zurdo-Pineiro JL, Mateos PF, Molina EM . (2004). Identification of microorganisms by PCR amplification and sequencing of a universal amplified ribosomal region present in both prokaryotes and eukaryotes. J Microbiol Methods 56: 413–426.
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD et al (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290.
Roh SW, Nam YD, Chang HW, Kim KH, Lee HJ, Oh HM et al (2007a). Natronococcus jeotgali sp. nov., a halophilic archaeon isolated from shrimp jeotgal, a traditional fermented seafood from Korea Int. J Syst Evol Microbiol 57: 2129–2131.
Roh SW, Nam YD, Chang HW, Sung Y, Kim KH, Oh HM et al (2007b). Halalkalicoccus jeotgali sp. nov., a halophilic archaeon from shrimp jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol 57: 2296–2298.
Roh SW, Sung Y, Nam YD, Chang HW, Kim KH, Yoon JH et al (2008). Arthrobacter soli sp. nov., a novel bacterium isolated from wastewater reservoir sediment. J Microbiol 46: 40–44.
Sands A, Crisan EV . (1974). Microflora of fermented Korean seafoods. J Food Sci 39: 1002–1005.
Schloss PD, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Schloss PD, Handelsman J . (2006). Toward a census of bacteria in soil. PLoS Comput Biol 2: e92.
Shen JP, Zhang LM, Zhu YG, Zhang JB, He JZ . (2008). Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10: 1601–1611.
Siboni N, Ben-Dov E, Sivan A, Kushmaro A . (2008). Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ Microbiol 10: 2979–2990.
Stackebrandt E, Goebel BM . (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44: 846–849.
Staley JT, Konopka A . (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39: 321–346.
Suh H-K, Yoon S-S . (1987). A study on the regional characteristics of Korean chotkal. Korean J Dietary Cult 2: 45–54.
Teske A, Sorensen KB . (2008). Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2: 3–18.
van Beek S, Priest FG . (2002). Evolution of the lactic acid bacterial community during malt whisky fermentation: a polyphasic study. Appl Environ Microbiol 68: 297–305.
Watanabe K, Kodama Y, Harayama S . (2001). Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods 44: 253–262.
Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P et al (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103: 12317–12322.
Yeates C, Gillings MR, Davison AD, Altavilla N, Veal DA . (1998). Methods for microbial DNA extraction from soil for PCR amplification. Biol Proced Online 1: 40–47.
Acknowledgements
This work was supported by the Environmental Biotechnology National Core Research Center (KOSEF: R15-2003-012-02002-0) and TDPAF (Technology Development Program for Agriculture and Forestry).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Roh, S., Kim, KH., Nam, YD. et al. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J 4, 1–16 (2010). https://doi.org/10.1038/ismej.2009.83
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.83
Keywords
This article is cited by
-
Terasi, exploring the Indonesian ethnic fermented shrimp paste
Journal of Ethnic Foods (2024)
-
Assessment of diversity of archaeal communities in Algerian chott
Extremophiles (2023)
-
Modeling microbial communities from atrazine contaminated soils promotes the development of biostimulation solutions
The ISME Journal (2019)
-
Changes in the metabolic potential of the sponge microbiome under ocean acidification
Nature Communications (2019)
-
Community structures and genomic features of undesirable white colony-forming yeasts on fermented vegetables
Journal of Microbiology (2019)