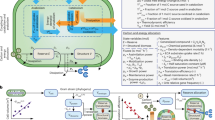
Abstract
The advent of rapid complete genome sequencing, and the potential to capture this information in genome-scale metabolic models, provide the possibility of comprehensively modeling microbial community interactions. For example, Rhodoferax and Geobacter species are acetate-oxidizing Fe(III)-reducers that compete in anoxic subsurface environments and this competition may have an influence on the in situ bioremediation of uranium-contaminated groundwater. Therefore, genome-scale models of Geobacter sulfurreducens and Rhodoferax ferrireducens were used to evaluate how Geobacter and Rhodoferax species might compete under diverse conditions found in a uranium-contaminated aquifer in Rifle, CO. The model predicted that at the low rates of acetate flux expected under natural conditions at the site, Rhodoferax will outcompete Geobacter as long as sufficient ammonium is available. The model also predicted that when high concentrations of acetate are added during in situ bioremediation, Geobacter species would predominate, consistent with field-scale observations. This can be attributed to the higher expected growth yields of Rhodoferax and the ability of Geobacter to fix nitrogen. The modeling predicted relative proportions of Geobacter and Rhodoferax in geochemically distinct zones of the Rifle site that were comparable to those that were previously documented with molecular techniques. The model also predicted that under nitrogen fixation, higher carbon and electron fluxes would be diverted toward respiration rather than biomass formation in Geobacter, providing a potential explanation for enhanced in situ U(VI) reduction in low-ammonium zones. These results show that genome-scale modeling can be a useful tool for predicting microbial interactions in subsurface environments and shows promise for designing bioremediation strategies.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R et al. (2003). Stimulating the In Situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol 69: 5884–5891.
Anesiadis N, Cluett WR, Mahadevan R . (2008). Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng 10: 255–266.
Balba MT, Nedwell DB . (1982). Microbial metabolism of acetate, propionate and butyrate in anoxic sediment from the Colne Point Saltmarsh, Essex, UK. J Gen Microbiol 128: 1415–1422.
Burgard AP, Pharkya P, Maranas CD . (2003). Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84: 647–657.
Caccavo F, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ . (1994). Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol 60: 3752–3759.
Chang YJ, Long PE, Geyer R, Peacock AD, Resch CT, Sublette K et al. (2005). Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ Sci Technol 39: 9039–9048.
Chapelle FH, Lovley DR . (1990). Rates of microbial metabolism in deep coastal plain aquifers. Appl Environ Microbiol 56: 1865–1874.
Crill PM, Martens CS . (1986). Methane production from bicarbonate and acetate in an anoxic marine sediment. Geochim Cosmochim Acta 50: 2089–2097.
Feist AM, Henry CS, Reed JL, Krummenacker M . (2007). A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121.
Feist AM, Palsson B . (2008). The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol 26: 659–667.
Fell DA, Small JR . (1986). Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem J 238: 781–786.
Finneran KT, Johnsen CV, Lovley DR . (2003). Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int J Syst Evol Microbiol 53: 669–673.
Hansen LK, Jakobsen R, Postma D . (2001). Methanogenesis in a shallow sandy aquifer, Rømø, Denmark. Geochimica et Cosmochimica Acta 65: 2925–2935.
Hjersted JL, Henson MA, Mahadevan R . (2007). Genome-scale analysis of Saccharomyces cerevisiae metabolism and ethanol production in fed-batch culture. Biotechnol Bioeng 97: 1190–1204.
Holmes DE, Bond DR, O’Neil RA, Reimers CE, Tender LR, Lovley DR . (2004). Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb Ecol 48: 178–190.
Holmes DE, Finneran KT, O’Neil RA, Lovley DR . (2002). Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl Environ Microbiol 68: 2300–2306.
Holmes DE, O’Neil RA, Chavan MA, N’Guessan LA, Vrionis HA, Perpetua LA et al. (2009). Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J 3: 216–230.
Holmes DE, O’Neil R, Vrionis HA, N’guessan L, Ortiz-Bernad I, Larrahondo MJ et al. (2007). Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J 1: 663–677.
Ibarra RU, Edwards JS, Palsson BO . (2002). Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420: 186–189.
Izallalen M, Mahadevan R, Burgard A, Postier B, Didonato R, Sun J et al. (2008). Geobacter sulfurreducens strain engineered for increased rates of respiration. Metab Eng 10: 267–275.
Komlos J, Peacock A, Kukkadapu RK, Jaffé PR . (2008). Long-term dynamics of uranium reduction/reoxidation under low sulfate conditions. Geochim Cosmochim Acta 72: 3603–3615.
Kuivila KM, Murray JW, Devol AH, Novelli PC . (1989). Methane production, sulfate reduction and competition for substrates in the sediments of Lake Washington. Geochim Cosmochim Acta 53: 409–416.
Lin B, Braster M, Röling WFM, van Breukelen BM . (2007). Iron-Reducing Microorganisms in a Landfill Leachate-Polluted Aquifer: Complementing Culture-Independent Information with Enrichments and Isolations. Geomicrobiol J 24: 283–294.
Lovley DR . (1991). Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev 55: 259–287.
Lovley DR . (2001). Bioremediation. Anaerobes to the rescue Science 293: 1444–1446.
Lovley DR . (2003). Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol 1: 35–44.
Lovley DR . (2006). Dissimilatory Fe(III)- and Mn(IV)-Reducing Prokaryotes. In: (eds). The Prokaryotes. Springer: New York, pp 1143.
Lovley DR, Chapelle FH . (1995). Deep subsurface microbial processes. Rev Geophys 33: 365–381.
Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, Gorby YA et al. (1993). Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol 159: 336–344.
Lovley DR, Holmes DE, Nevin KP . (2004). Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol 49: 219–286.
Lovley DR, Klug MJ . (1983). Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl Environ Microbiol 45: 187–192.
Lovley DR, Klug MJ . (1986). Model for the distribution of sulfate reduction and methanogenesis in freshwater sediments. Geochim Cosmochim Acta 50: 11–18.
Lovley DR, Phillips EJ . (1989). Requirement for a microbial consortium to completely oxidize glucose in Fe(III)-reducing sediments. Appl Environ Microbiol 55: 3234–3236.
Mahadevan R, Bond DR, Butler JE, Esteve-Nuñez A, Coppi MV, Palsson BO et al. (2006). Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol 72: 1558–1568.
Mahadevan R, Edwards JS, Doyle FJ . (2002). Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J 83: 1331–1340.
Methe BA, Nelson KE, Eisen JA, Paulsen IT . (2003). Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302: 1967–1969.
Methe BA, Webster J, Nevin K, Butler J, Lovley DR . (2005). DNA microarray analysis of nitrogen fixation and Fe(III) reduction in Geobacter sulfurreducens. Appl Environ Microbiol 71: 2530–2538.
Mouser PJ, N’Guessan AL, Elifantz H, Holmes DE, Williams KH, Wilkins MJ et al. (2009). Influence of heterogeneous ammonium availability on bacterial community structure and the expression of nitrogen fixation and ammonium transporter genes during in situ bioremediation of uranium-contaminated groundwater. Environ Sci Technol 43: 4386–4392.
Oberhardt MA, Palsson BO, Papin JA . (2009). Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5: 320.
Petrie L, North NN, Dollhopf SL, Balkwill DL, Kostka JE . (2003). Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl Environ Microbiol 69: 7467–7479.
Pfeiffer T, Bonhoeffer S . (2003). An evolutionary scenario for the transition to undifferentiated multicellularity. Proceedings of the National Academy of Sciences 100: 1095–1098.
Pfeiffer T, Bonhoeffer S . (2004). Evolution of cross—feeding in microbial populations. Am Nat 163: E126–E135.
Pfeiffer T, Schuster S . (2005). Game-theoretical approaches to studying the evolution of biochemical systems. Trends Biochem Sci 30: 20–25.
Pfeiffer T, Schuster S, Bonhoeffer S . (2001). Cooperation and competition in the evolution of ATP-producing pathways. Science 292: 504–507.
Pharkya P, Burgard AP, Maranas CD . (2003). Exploring the overproduction of amino acids using the bilevel optimization framework OptKnock. Biotechnol Bioeng 84: 887–899.
Pharkya P, Burgard AP, Maranas CD . (2004). OptStrain: a computational framework for redesign of microbial production systems. Genome Res 14: 2367–2376.
Pharkya P, Maranas CD . (2006). An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng 8: 1–13.
Reed JL, Palsson B . (2003). Thirteen years of building constraint-based in silico models of Escherichia coli. J Bacteriol 185: 2692–2699.
Reed JL, Vo TD, Schilling CH, Palsson B . (2003). An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54.
Risso C, Sun J, Zhuang K, Mahadevan R, DeBoy R, Ismail W et al. (2009). Genome-scale comparison and constraint-based metabolic reconstruction of the facultative anaerobic Fe(III)-reducer Rhodoferax ferrireducens. BMC Genomics 10: 447.
Scheibe TD, Mahadevan R, Fang Y, Garg S . (2009). Coupling a genome-scale metabolic model with a reactive transport model to describe in situ uranium bioremediation. Microb Biotechnol 2: 274–286.
Schuster S, Pfeiffer T, Fell DA . (2008). Is maximization of molar yield in metabolic networks favoured by evolution? J Theor Biol 252: 497–504.
Segura D, Mahadevan R, Juárez K, Lovley DR . (2008). Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PLoS Comput Biol 4: e36.
Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA et al. (2007). Metabolic modeling of a mutualistic microbial community. Mol Syst Biol 3: 92.
Strycharz A, Peacock A, Williams KH, N’Guessan L, Lovley DR . (2009). Geobacter species are predominately planktonic during growth in uranium-contaminated subsurface sediments. American Society for Microbiolgy 109th General Meeting 14: 447.
Sun J, Sayyar B, Butler JE, Pharkya P, Fahland TR, Famili I et al. (2009). Genome-scale constraint-based modeling of Geobacter metallireducens. BMC Syst Biol 3: 15.
Varma A, Palsson BO . (1994). Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol 60: 3724–3731.
Vrionis HA, Anderson RT, Ortiz-Bernad I, Ortiz-Bernad I, O’Neil KR, Resch CT et al. (2005). Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol 71: 6308–6318.
Wall JD, Krumholz LR . (2006). Uranium reduction. Annu Rev Microbiol 60: 149–166.
Watson MR . (1986). A discrete model of bacterial metabolism. Comput Appl Biosci 2: 23–27.
Wilkins MJ, Verberkmoes NC, Williams KH, Callister SJ, Mouser PJ, Elifantz H et al. (2009). Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl Environ Microbiol 75: 6591–6599.
Yabusaki SB, Fang Y, Long PE, Resch CT, Peacock AD, Komlos J et al. (2007). Uranium removal from groundwater via in situ biostimulation: field-scale modeling of transport and biological processes. J Contam Hydrol 93: 216–235.
Acknowledgements
This research was supported by the Office of Science (BER), the US Department of Energy, Cooperative Agreement No. DE-FC02-02ER63446 and Grant No. DE-FG02-07ER64367; this research was also supported by the Canada Foundation for Innovation and the University of Toronto Open Fellowship, as well as the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405). We thank Caitlin O’Connel for her artistic illustration, Laurence Yang for his insightful discussions and Emma Janssen for her grammatical corrections.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Appendix
Rights and permissions
About this article
Cite this article
Zhuang, K., Izallalen, M., Mouser, P. et al. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J 5, 305–316 (2011). https://doi.org/10.1038/ismej.2010.117
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.117
Keywords
This article is cited by
-
Effect of silver nanoparticles on nitrogen-cycling bacteria in constructed wetlands
Nanotechnology for Environmental Engineering (2022)
-
Redox-informed models of global biogeochemical cycles
Nature Communications (2020)
-
Extracellular electron uptake in Methanosarcinales is independent of multiheme c-type cytochromes
Scientific Reports (2020)
-
Genome-scale reconstruction of Paenarthrobacter aurescens TC1 metabolic model towards the study of atrazine bioremediation
Scientific Reports (2020)
-
Genome-scale Modeling of Metabolism and Macromolecular Expression and Their Applications
Biotechnology and Bioprocess Engineering (2020)