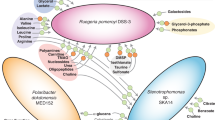
Abstract
Metaproteomics is one of a suite of new approaches providing insights into the activities of microorganisms in natural environments. Proteins, the final products of gene expression, indicate cellular priorities, taking into account both transcriptional and posttranscriptional control mechanisms that control adaptive responses. Here, we report the proteomic composition of the < 1.2 μm fraction of a microbial community from Oregon coast summer surface waters, detected with two-dimensional liquid chromatography coupled with electrospray tandem mass spectrometry. Spectra corresponding to proteins involved in protein folding and biosynthesis, transport, and viral capsid structure were the most frequently detected. A total of 36% of all the detected proteins were best matches to the SAR11 clade, and other abundant coastal microbial clades were also well represented, including the Roseobacter clade (17%), oligotrophic marine gammaproteobacteria group (6%), OM43 clade (1%). Viral origins were attributed to 2.5% of proteins. In contrast to oligotrophic waters, phosphate transporters were not highly detected in this nutrient-rich system. However, transporters for amino acids, taurine, polyamines and glutamine synthetase were among the most highly detected proteins, supporting predictions that carbon and nitrogen are more limiting than phosphate in this environment. Intriguingly, one of the highly detected proteins was methanol dehydrogenase originating from the OM43 clade, providing further support for recent reports that the metabolism of one-carbon compounds by these streamlined methylotrophs might be an important feature of coastal ocean biogeochemistry.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Allgaier M, Uphoff H, Felske A, Wagner-Dobler I . (2003). Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl Environ Microbiol 69: 5051–5059.
Beja O, Suzuki MT, Heidelberg JF, Nelson KE, Preston CM, Hamada T et al. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415: 630–633.
Bouvier T, del Giorgio PA . (2007). Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ Microbiol 9: 287–297.
Brinkoff T, Giebel H-A, Simon M . (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189: 531–539.
Button DK, Robertson B . (2000). Effect of nutrient kinetics and cytoarchitecture on bacterioplankter size. Limnol Oceanogr 45: 499–505.
Cho JC, Giovannoni SJ . (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol 70: 432–440.
Cho JC, Stapels MD, Morris RM, Vergin KL, Schwalbach MS, Givan SA et al. (2007). Polyphyletic photosynthetic reaction centre genes in oligotrophic marine Gammaproteobacteria. Environ Microbiol 9: 1456–1463.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
Dyhrman ST, Haley ST . (2006). Phosphorus scavenging in the unicellular marine diazotroph Crocosphaera watsonii. Appl Environ Microbiol 72: 1452–1458.
Eng J, McCormack AL, Yates III JR . (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989.
Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810.
Fuchs BM, Spring S, Teeling H, Quast C, Wulf J, Schattenhofer M et al. (2007). Characterization of a marine gammaproteobacterium capable of aerobic anoxygenic photosynthesis. Proc Natl Acad Sci USA 104: 2891–2896.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL et al. (2005a). Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005b). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Goericke R . (2002). Bacteriochlorophyll a in the ocean: Is anoxygenic bacterial photosynthesis important? Limnol Oceanogr 47: 290–295.
Hoch MP, Jeffrey WH, Snyder RA, Dillon KS, Coffin RB . (2006). Expression of glutamine synthetase and glutamate dehydrogenase by marine bacterioplankton: assay optimization and efficacy for assessing nitrogen to carbon metabolic balance in situ. Limnol Oceanogr 4: 308–328.
Kan J, Hanson TE, Ginter JM, Wang K, Chen F . (2005). Metaproteomic analysis of Chesapeake Bay microbial communities. Saline Systems 1: 7.
Karp-Boss L, Wheeler PA, Hales B, Covert P . (2004). Distributions and variability of particulate organic matter in a coastal upwelling system. J Geophys Res 109: C09010.
Lee S, Fuhrman JA . (1987). Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol 53: 1298–1303.
Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, DiBartolo G. et al. (2007). Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446: 537–541.
Martiny AC, Coleman ML, Chisholm SW . (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA 103: 12552–12557.
McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR . (2002). Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. 219: 245–251.
Miller TR, Belas R . (2004). Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl Environ Microbiol 70: 3383–3391.
Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913.
Moran MA, Gonzalez JM, Kiene RP . (2003). Linking a Bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol J 20: 375–388.
Morris RM, Longnecker K, Giovannoni SJ . (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8: 1361–1370.
Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G . (2010). Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J 4: 673–685.
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Muller-Karger FE, Varela R, Thunell R, Luerssen R, Hu C, Walsh JJ . (2005). The importance of continental margins in the global carbon cycle. Geophys Res Lett 32: L01602.
Park K . (1967). Nutrient regeneration and preformed nutrients off oregon. Limnol Oceanogr 12: 353–357.
Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP . (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2: 43–50.
Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M et al. (2005). Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol 71: 4121–4126.
Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC et al. (2005). Community proteomics of a natural microbial biofilm. Science 308: 1915–1920.
Rappe MS, Vergin K, Giovannoni SJ . (2000). Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol 33: 219–232.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77.
Scanlan DJ, Silman NJ, Donald KM, Wilson WH, Carr NG, Joint I et al. (1997). An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol 63: 2411–2420.
Sherr EB, Sherr BF, Wheeler PA . (2005). Distribution of coccoid cyanobacteria and small eukaryotic phytoplankton in the upwelling ecosystem off the Oregon coast during 2001 and 2002. Deep Sea Res Part 2 Top Stud Oceanogr 52: 317–330.
Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky D et al. (2008). Transport functions dominate the SAR11 metaproteome at low nutrient extremes in the Sargasso Sea. ISME J 3: 93–105.
Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH . (2001). Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep-Sea Research II 48: 1405–1447.
Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ . (2007). The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol 73: 2290–2296.
Tabb DL, McDonald WH, Yates III JR . (2002). DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26.
Thompson MR, Chourey K, Froelich JM, Erickson BK, VerBerkmoes NC, Hettich RL . (2008). Experimental approach for deep proteome measurements from small-scale microbial biomass samples. 80: 9517–9525.
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al. (2005). Comparative metagenomics of microbial communities. Science 308: 554–557.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J et al. (2008). Shotgun metaproteomics of the human distal gut microbiota. ISME J 3: 179–189.
Wheeler PA, Huyer A, Fleischbein J . (2003). Cold halocline, increased nutrients and higher chlorophyll off Oregon in 2002. Geophys Res Lett (online) 30: 8021; doi:10.1029/2003GL017395.
Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ . (2007). Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct 2: 27.
Wu J, Sunda W, Boyle EA, Karl DM . (2000). Phosphate depletion in the western North Atlantic Ocean. Science 289: 759–762.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K et al. (2007). The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol 5: e16.
Zubkov MV, Sleigh MA, Burkill PH . (2000). Assaying picoplankton distribution by flow cytometry of underway samples collected along a meridional transect across the Atlantic Ocean. Aquat Microb Ecol 21: 13–20.
Acknowledgements
We thank the crew of the Elaka and Joshua Kitner for their help in sample collection, and Francis Chan for data and discussions about oceanographic conditions. This work was supported in part by a Marine Microbiology Initiative Investigator Award from the Gordon and Betty Moore Foundation. Additional sponsorship was received from the US Department of Energy under contract DE-AC05-00OR22725 with Oak Ridge National Laboratory, managed and operated by UT-Battelle, LLC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Sowell, S., Abraham, P., Shah, M. et al. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5, 856–865 (2011). https://doi.org/10.1038/ismej.2010.168
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.168
Keywords
This article is cited by
-
Compartment-related aspects of XoxF protein functionality in Methylorubrum extorquens DM4 analysed using its cytoplasmic targeting
Antonie van Leeuwenhoek (2023)
-
Metabolic tuning of a stable microbial community in the surface oligotrophic Indian Ocean revealed by integrated meta-omics
Marine Life Science & Technology (2022)
-
Protist diversity and community complexity in the rhizosphere of switchgrass are dynamic as plants develop
Microbiome (2021)
-
Bacterial community variations in the South China Sea driven by different chemical conditions
Ecotoxicology (2021)
-
Metaproteomics reveal that rapid perturbations in organic matter prioritize functional restructuring over taxonomy in western Arctic Ocean microbiomes
The ISME Journal (2020)