Abstract
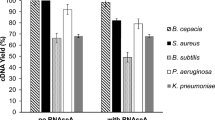
Metatranscriptomes generated by pyrosequencing hold significant potential for describing functional processes in complex microbial communities. Meeting this potential requires protocols that maximize mRNA recovery by reducing the relative abundance of ribosomal RNA, as well as systematic comparisons to identify methodological artifacts and test for reproducibility across data sets. Here, we implement a protocol for subtractive hybridization of bacterial rRNA (16S and 23S) that uses sample-specific probes and is applicable across diverse environmental samples. To test this method, rRNA-subtracted and unsubtracted transcriptomes were sequenced (454 FLX technology) from bacterioplankton communities at two depths in the oligotrophic open ocean, yielding 10 data sets representing ∼350 Mbp. Subtractive hybridization reduced bacterial rRNA transcript abundance by 40–58%, increasing recovery of non-rRNA sequences up to fourfold (from 12% to 20% of total sequences to 40–49%). In testing this method, we established criteria for detecting sequences replicated artificially via pyrosequencing errors and identified such replicates as a significant component (6–39%) of total pyrosequencing reads. Following replicate removal, statistical comparisons of reference genes (identified via BLASTX to NCBI-nr) between technical replicates and between rRNA-subtracted and unsubtracted samples showed low levels of differential transcript abundance (<0.2% of reference genes). However, gene overlap between data sets was remarkably low, with no two data sets (including duplicate runs from the same pyrosequencing library template) sharing greater than 17% of unique reference genes. These results indicate that pyrosequencing captures a small subset of total mRNA diversity and underscores the importance of reliable rRNA subtraction procedures to enhance sequencing coverage across the functional transcript pool.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S et al. (2009). Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 6: 647–649.
Audic S, Claverie JM . (1997). The significance of digital gene expression profiles. Genome Res 7: 986–995.
Ballantyne KN, van Oorschot RAH, Muharam I, van Daal A, Mitchell RJ . (2007). Decreasing amplification bias associated with multiple displacement amplification and short tandem repeat genotyping. Anal Biochem 368: 222–229.
Benjamini Y, Hochberg Y . (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300.
Bergen AW, Qi Y, Haque KA, Welch RA, Chanock SJ . (2005). Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. BMC Biotechnol 5: 24.
Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, Prüfer K et al. (2007). Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA 104: 14616–14621.
Dalevi D, Ivanova NN, Mavromatis K, Hooper SD, Szeto E, Hugenholtz P et al. (2008). Annotation of metagenome short reads using proxygenes. Bioinformatics 24: i7–i13.
DeLong EF . (2009). The microbial ocean from genomes to biomes. Nature 459: 200–206.
DeLong EF, Taylor LT, Marsh TL, Preston CM . (1999). Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol 65: 5554–5563.
Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810.
Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P et al. (2008). Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3: e3042.
Gomez-Alvarez V, Teal TK, Schmidt TM . (2009). Systematic artifacts in metagenomes from complex microbial communities. ISMEJ 3: 1–4.
Hewson I, Poretsky RS, Beinart RA, White AE, Shi T, Bench SR et al. (2009a). In situ transcriptomic analysis of the globally important keystone N2-fixing taxon Crocosphaera watsonii. ISMEJ 3: 618–631.
Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ et al. (2009b). Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISMEJ, 3: 1286–1300.
Hunt DE, Klepac-Ceraj V, Acinas SG, Gautier C, Bertilsson S, Polz MF . (2006). Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl Environ Microbiol 72: 2221–2225.
Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC . (2009). Methods for comparative metagenomics. BMC Bioinformatics 10 (Suppl 1): S12.
Li W, Godzik A . (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659.
McGrath KC, Thomas-Hall SR, Cheng CT, Leo L, Alexa A, Schmidt S et al. (2008). Isolation and analysis of mRNA from environmental microbial communities. J Microbiol Meth 75: 172–176.
Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M et al. (2005). Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol 71: 4121–4126.
Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA . (2009). Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol 11: 1359–1375.
Rodriguez-Brito B, Rohwer F, Edwards RA . (2006). An application of statistics to comparative metagenomics. BMC Bioinformatics 7: 162.
Shi Y, Tyson GW, DeLong EF . (2009). Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459: 266–269.
Su C, Sordillo LM . (1998). A simple method to enrich mRNA from total prokaryotic RNA. Mol Biotechnol 10: 83–85.
Urich T, Lamzen A, Qi J, Huson DH, Schleper C, Schuster SC . (2008). Simultaneous assessment of soil microbial community structure and function through analysis of the metatranscriptome. PLoS One 3: e2527.
Wendisch VF, Zimmer DP, Khodursky A, Peter B, Cozzarelli N, Kustu S . (2001). Isolation of Escherichia coli mRNA and comparison using mRNA and total RNA on DNA microarrays. Anal Biochem 290: 205–213.
Yoder-Himes DR, Chain PSG, Zhu Y, Wurtzel O, Rubin EM, Tiedje JM et al. (2009). Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci USA 106: 3976–3981.
Acknowledgements
We thank Yanmei Shi and the captain and crew of the R/V Kilo Moana for their help in collecting samples for this study, Jay McCarren and Asuncion Martinez for help with DNA extractions, Rachel Barry for her tireless work in preparing samples for pyrosequencing, Tracy Teal for providing scripts for use in identifying replicate pyrosequencing reads, Hiro Kimura for generously providing data on rRNA subtraction efficiency in Dokdonia, and Adrian Sharma for providing data on rRNA subtraction efficiency in marine bacterioplankton samples from Bermuda. This work was supported by a gift from the Agouron Institute, and grants from the Gordon and Betty Moore Foundation (EFD), the Office of Science (BER) US Department of Energy, and NSF Science and Technology Center Award EF0424599. This work is a contribution of the Center for Microbial Oceanography: Research and Education (C-MORE)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Stewart, F., Ottesen, E. & DeLong, E. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4, 896–907 (2010). https://doi.org/10.1038/ismej.2010.18
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.18
Keywords
This article is cited by
-
Soil microbial ecology through the lens of metatranscriptomics
Soil Ecology Letters (2024)
-
MetaPro: a scalable and reproducible data processing and analysis pipeline for metatranscriptomic investigation of microbial communities
Microbiome (2023)
-
Blocking Abundant RNA Transcripts by High-Affinity Oligonucleotides during Transcriptome Library Preparation
Biological Procedures Online (2023)
-
Comparison of rRNA depletion methods for efficient bacterial mRNA sequencing
Scientific Reports (2022)
-
Wounding response in Porifera (sponges) activates ancestral signaling cascades involved in animal healing, regeneration, and cancer
Scientific Reports (2022)