Abstract
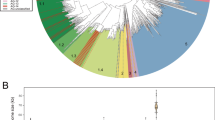
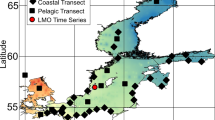
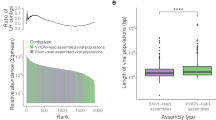
Knowledge of marine phages is highly biased toward double-stranded DNA (dsDNA) phages; however, recent metagenomic surveys have also identified single-stranded DNA (ssDNA) phages in the oceans. Here, we describe two complete ssDNA phage genomes that were reconstructed from a viral metagenome from 80 m depth at the Bermuda Atlantic Time-series Study (BATS) site in the northwestern Sargasso Sea and examine their spatial and temporal distributions. Both genomes (SARssφ1 and SARssφ2) exhibited similarity to known phages of the Microviridae family in terms of size, GC content, genome organization and protein sequence. PCR amplification of the replication initiation protein (Rep) gene revealed narrow and distinct depth distributions for the newly described ssDNA phages within the upper 200 m of the water column at the BATS site. Comparison of Rep gene sequences obtained from the BATS site over time revealed changes in the diversity of ssDNA phages over monthly time scales, although some nearly identical sequences were recovered from samples collected 4 years apart. Examination of ssDNA phage diversity along transects through the North Atlantic Ocean revealed a positive correlation between genetic distance and geographic distance between sampling sites. Together, the data suggest fundamental differences between the distribution of these ssDNA phages and the distribution of known marine dsDNA phages, possibly because of differences in host range, host distribution, virion stability, or viral evolution mechanisms and rates. Future work needs to elucidate the host ranges for oceanic ssDNA phages and determine their ecological roles in the marine ecosystem.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Abascal F, Zardoya R, Posada D . (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Angly F, Felts B, Breitbart M, Salamon P, Edwards R, Carlson C et al. (2006). The marine viromes of four oceanic regions. PLoS Biol 4: e368.
Anisimova M, Gascuel O . (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552.
Breitbart M, Felts B, Kelley S, Mahaffy JM, Nulton J, Salamon P et al. (2004). Diversity and population structure of a near-shore marine-sediment viral community. Proc R Soc Lond B Biol Sci 271: 565–574.
Breitbart M, Rohwer F . (2004). Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett 236: 245–252.
Breitbart M, Rohwer F . (2005). Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13: 278–284.
Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D et al. (2002). Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA 99: 14250–14255.
Brentlinger K, Hafenstein S, Novak C, Fane B, Borgon R, McKenna R et al. (2002). Microviridae, a family divided: isolation, characterization, and genome sequence of φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J Bacteriol 184: 1089–1094.
Carlson C, Morris R, Parsons R, Treusch A, Giovannoni S, Vergin K . (2009). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295.
Cuevas J, Duffy S, Sanjuan R . (2009). Point mutation rate of bacteriophage φX174. Genetics 183: 747–749.
Culley A, Steward G . (2007). New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl Environ Microbiol 73: 5937–5944.
Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M et al. (2008). Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452: 340–343.
Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452: 629–632.
Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Heled J et al. (2010). Geneious v5.0.4 http://www.geneious.com.
Drummond A, Strimmer K . (2001). PAL: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics 17: 662–663.
Duffy S, Holmes E . (2009). Validation of high rates of nucleotide substitution in geminiviruses: phylogenetic evidence from East African cassava mosaic viruses. J Gen Virol 90: 1539–1547.
Duffy S, Shackelton L, Holmes E . (2008). Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9: 267–276.
Edgar R . (2004a). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Edgar R . (2004b). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5: 113.
Fane B (2005). Family microviridae. In: Fauquet C, Mayo M, Maniloff J, Desselberger U, Ball L (eds). Virus Taxonomy: Classification and Nomenclature of Viruses. Elsevier Academic Press: San Diego, pp 289–299.
Faruque SM, Rahman MM, Asadulghani, Islam KMN, Mekalanos JJ . (1999). Lysogenic conversion of environmental Vibrio mimicus strains by CTXφ. Infect Immun 67: 5723–5729.
Fauquet C, Mayo M, Maniloff J, Desselberger U, Ball L (eds) (2005). Virus Taxonomy: Classification and Nomenclature of Viruses. Elsevier Academic Press: San Diego.
Fuhrman J, Schwalbach M . (2003). Viral influence of aquatic bacterial communities. Biol Bull 204: 192–195.
Garner S, Everson J, Lambden P, Fane B, Clarke I . (2004). Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila percorum. Virus Genes 28: 207–214.
Guindon S, Daufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O . (2010). New algorithms and methods to estimate maximum likelihood phylogenies: assessing the performance of PhyML3.0. Syst Biol 59: 307–321.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Haible D, Kober S, Jeske H . (2006). Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J Virol Methods 135: 9–16.
Hall T . (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98.
Huang SJ, Wilhelm SW, Jiao NAZ, Chen F . (2010). Ubiquitous cyanobacterial podoviruses in the global oceans unveiled through viral DNA polymerase gene sequences. ISME J 4: 1243–1251.
Jiang SC, Paul JH . (1998). Gene transfer by transduction in the marine environment. Appl Environ Microbiol 64: 2780–2787.
Kellogg CA, Rose JB, Jiang SC, Turmond JM, Paul JH . (1995). Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar Ecol Prog Ser 120: 89–98.
Kim K, Chang H, Nam Y, Roh S, Kim M, Sung Y et al. (2008). Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol 74: 5975–5985.
Labonte JM, Reid KE, Suttle CA . (2009). Phylogenetic analysis indicates evolutionary diversity and environmental segregation of marine podovirus DNA polymerase gene sequences. Appl Environ Microbiol 75: 3634–3640.
LeClerc J . (2002). Single-stranded DNA phages. In: Streips U, Yasbin R (eds). Modern Microbial Genetics 2nd edn. Wiley-Liss, Inc.: New York, pp 145–176.
Lui B, Everson J, Fane B, Giannikopoulou P, Vertou E, Lambden P et al. (2000). Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J Virol 74: 3464–3469.
Marie D, Brussaard C, Thyrhaug R, Bratbak G, Vaulot D . (1999). Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol 65: 45–52.
McDaniel L, Paul J, de la Rosa M . (2006). Temperate and lytic cyanophages from the Gulf of Mexico. J Mar Biol Assoc UK 86: 517–527.
Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ . (2005). Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50: 1687–1696.
Nagasaki K, Tomaru Y, Takao Y, Nishida K, Shirai Y, Suzuki H et al. (2005). Previously unknown virus infects marine diatom. Appl Environ Microbiol 71: 3528–3535.
Parks D, Porter M, Churcher S, Wang S, Blouin C, Whalley J et al. (2009). GenGIS: a geospatial information system for genomic data. Genome Res 19: 1896–1904.
Paul JH . (2008). Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J 2: 579–589.
Posada D . (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256.
Read T, Brunham R, Shen C, Gill S, Heidelberg J, White O et al. (2000). Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28: 1397–1406.
Renaudin J, Paracel M, Bove J . (1987). Spiroplasma virus 4: nucleotide sequence of the viral DNA, regulatory signals, and the proposed genome organization. J Bacteriol 169: 4950–4961.
Rohwer F, Thurber RV . (2009). Viruses manipulate the marine environment. Nature 459: 207–212.
Rokyta D, Burch C, Caudle S, Wichman H . (2006). Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol 188: 1134–1142.
Rosario K, Duffy S, Breitbart M . (2009). Diverse circovirus-like genome architectures revealed by environmental metagenomics. J Gen Virol 90: 2418–2424.
Rozen S, Skaletsky H . (2000). Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds). Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press: Totowa, NJ, pp 365–386.
Sambrook J, Fritsch E, Maniatis T . (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press: New York.
Sandaa R . (2008). Burden or benefit? Virus–host interactions in the marine environment. Res Microbiol 159: 374–381.
Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA et al. (1977). Nucleotide sequence of bacteriophage φX174 DNA. Nature 265: 687–695.
Short C, Suttle C . (2005). Nearly identical bacteriophage structural gene sequences are widely distributed in marine and freshwater environments. Appl Environ Microbiol 71: 480–486.
Short SM, Suttle CA . (2002). Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl Environ Microbiol 68: 1290–1296.
Steward G . (2001). Fingerprinting viral assemblages by pulsed field gel electrophoresis. In: Paul J (ed). Methods in Microbiology. Academic Press: San Diego, pp 85–103.
Steward G, Montiel J, Azam F . (2000). Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol Oceanogr 45: 1697–1706.
Storey CC, Lusher M, Richmond SJ . (1989). Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J Gen Virol 70: 3381–3390.
Sullivan M, Coleman M, Quinlivan V, Rosenkrantz J, DeFrancesco A, Tan G et al. (2008). Portal protein diversity and phage ecology. Enviro Microbiol 10: 2810–2823.
Thurber R, Haynes M, Breitbart M, Wegley L, Rohwer F . (2009). Laboratory procedures to generate viral metagenomes. Nat Protocols 4: 470–483.
Tomaru Y, Nagasaki K . (2007). Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J Oceanogr 63: 215–221.
Tomaru Y, Shirai Y, Suzuki H, Nagumo T, Nagasaki K . (2008). Isolation and characterization of a new single-stranded DNA virus infecting the cosmopolitan marine diatom Chaetoceros debilis. Aqua Microbial Ecol 50: 103–112.
Treusch A, Vergin K, Finlay L, Donatz M, Burton R, Carlson C et al. (2009). Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3: 1148–1163.
Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F . (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9: 2707–2719.
Weinbauer M . (2004). Ecology of prokaryotic viruses. FEMS Microbiol Rev 28: 127–181.
Weinbauer M, Rassoulzadegan F . (2004). Are viruses driving microbial diversification and diversity? Environ Microbiol 6: 1–11.
Wommack K, Colwell R . (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64: 69–114.
Xia X, Xie Z . (2001). DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92: 371–373.
Yu Y, Breitbart M, McNairnie P, Rohwer F . (2006). FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinform 7: 57.
Acknowledgements
This research was funded by a grant to MB from the National Science Foundation (MCB-0701984). Thanks to Steven Bell, Craig Carlson, Dawn Goldsmith, Kevin Lew, Ben Van Mooy, Karyna Rosario, and the crew of the RV Atlantic Explorer for help collecting and processing samples. Thanks to Bhakti Dwivedi for bioinformatic assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Tucker, K., Parsons, R., Symonds, E. et al. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J 5, 822–830 (2011). https://doi.org/10.1038/ismej.2010.188
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.188
Keywords
This article is cited by
-
Expanding known viral diversity in plants: virome of 161 species alongside an ancient canal
Environmental Microbiome (2022)
-
VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences
Microbiome (2020)
-
An Insight into Phage Diversity at Environmental Habitats using Comparative Metagenomics Approach
Current Microbiology (2018)
-
Diversity and Distribution Characteristics of Viruses in Soils of a Marine-Terrestrial Ecotone in East China
Microbial Ecology (2018)
-
Genomic exploration of individual giant ocean viruses
The ISME Journal (2017)