Abstract
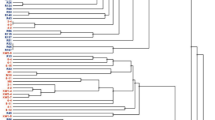
Our current knowledge of plant-microbe interactions indicate that populations inhabiting a host plant are not restricted to a single microbial species but comprise several genera and species. No one knows if communities inside plants interact, and it has been speculated that beneficial effects are the result of their combined activities. During an ecological study of nitrogen-fixing bacterial communities from Lupinus angustifolius collected in Spain, significant numbers of orange-pigmented actinomycete colonies were isolated from surface-sterilized root nodules. The isolates were analysed by BOX-PCR fingerprinting revealing an unexpectedly high genetic variation. Selected strains were chosen for 16S rRNA gene sequencing and phylogenetic analyses confirmed that all strains isolated belonged to the genus Micromonospora and that some of them may represent new species. To determine the possibility that the isolates fixed atmospheric nitrogen, chosen strains were grown in nitrogen-free media, obtaining in some cases, significant growth when compared with the controls. These strains were further screened for the presence of the nifH gene encoding dinitrogenase reductase, a key enzyme in nitrogen fixation. The partial nifH-like gene sequences obtained showed a 99% similarity with the sequence of the nifH gene from Frankia alni ACN14a, an actinobacterium that induces nodulation and fixes nitrogen in symbiosis with Alnus. In addition, in situ hybridization was performed to determine if these microorganisms inhabit the inside of the nodules. This study strongly suggests that Micromonospora populations are natural inhabitants of nitrogen-fixing root nodules.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Amann R, Ludwig W, Schleifer K . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Bray RH, Kurtz IT . (1945). Determination of total organic and available forms of phosphorous in soils. Soil Sci 59: 39–45.
Bremner JM, Mulvaney CS . (1982). Total Nitrogen. In: Page AL, Millar RH (eds). Methods of Soil Analysis Part 2, Chemical and Microbiological Properties. American Society of Agronomy: Madison. pp 621–622.
Carro L . (2009). Avances en la sistemática del género Micromonospora: estudio de cepas aisladas de la rizosfera y nódulos de Psium sativum. Ph.D. Thesis. Universidad de Salamanca, Spain.
Cerda E . (2008). Aislamiento de Micromonospora de nódulos de leguminosas tropicales y análisis de su interés como promotor del crecimiento vegetal. Ph.D. Thesis. Universidad de Salamanca, Spain.
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK et al. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57: 2259–2261.
Coombs JT, Franco CM . (2003). Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol 69: 5603–5608.
Coombs JT, Michelsen PP, Franco CM . (2004). Evaluation of endophytic actinobacteria as antagonists of Gaeumannomyces graminis var. tritici in wheat. Biol Control 29: 359–366.
de Maria N, de Felipe MR, Fernández-Pascual M . (2005). Alterations induced by glyphosate on lupin photosynthetic apparatus and nodule ultrastructure and some oxygen diffusion related proteins. Plant Physiol Biochem 43: 985–993.
de Menezes AB, Lockhart RJ, Cox MJ, Allison HE, McCarthy AJ . (2008). Cellulose degradation by micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA gyrase B gene sequencing. Appl Environ Microbiol 74: 7080–7084.
Doetsch RN . (1981). Determinative methods of light microscopy. In: Gerdhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds). Manual of Methods for General Bacteriology. American Society for Microbiology: Washington. pp 21–33.
Garcia LC, Martínez-Molina E, Trujillo ME . (2010). Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int J Syst Evol Microbiol 60: 331–337.
Gtari M, Brusetti L, Hassen A, Mora D, Daffoncihio D, Boudabous A . (2007). Genetic diversity among Elaeagnus compatible Frankia strains and sympatric-related nitrogen-fixing actinobacteria revealed by nifH sequence analysis. Soil Biol Biochem 39: 372–377.
Igarashi Y, Trujillo ME, Martinez-Molina E, Yanase S, Miyanaga S, Obata T et al. (2007). Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg Med Chem Lett 17: 3702–3705.
Ismet A, Vikineswarg S, Paramaswari S, Wang W, Ward A, Seki T et al. (2004). Production and chemical characterization of antifungal metabolites from Micromonospora sp. M39 isolated from mangrove rhizosphere soil. World J Microbiol Biotechnol 20: 523–528.
Jarabo-Lorenzo A, Perez-Galdona R, Donate-Correa J, Rivas R, Velazquez E, Hernandez M et al. (2003). Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst Appl Microbiol 26: 611–623.
Jongrungruangchok S, Tanasupawat S, Kudo T . (2008). Micromonospora chaiyaphumensis sp. nov., isolated from Thai soils. Int J Syst Evol Microbiol 58: 924–928.
Kasai H, Tamura T, Harayama S . (2000). Intrageneric relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int J Syst Evol Microbiol 50 (Part 1): 127–134.
Kawamoto I . (1989). Genus Micromonospora Ørskov 1923, 147AL. In: Williams ST, Sharpe ME, Holt JG (eds). Bergey's Manual of Systematic Bacteriology. Williams & Wilkins: Baltimore. pp 2442–2450.
Kimura M . (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Lane DJ . (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds). Nucleic acid Techniques in Bacterial Systematics. Wiley: New York. pp 115–175.
Lanoot B, Vancanneyt M, Dawyndt P, Cnockaert M, Zhang J, Huang Y et al. (2004). BOX-pCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst Appl Microbiol 27: 84–92.
Maldonado LA, Stach JE, Pathom-aree W, Ward AC, Bull AT, Goodfellow M . (2005). Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie Van Leeuwenhoek 87: 11–18.
Maldonado LA, Stach JE, Ward AC, Bull AT, Goodfellow M . (2008). Characterisation of micromonosporae from aquatic environments using molecular taxonomic methods. Antonie Van Leeuwenhoek 94: 289–298.
Normand P, Simonet P, Bardin R . (1988). Conservation of nif sequences in Frankia. Mol Gen Genet 213: 238–246.
Oke V, Long SR . (1999). Bacteroid formation in the Rhizobium–legume symbiosis. Curr Opin Microbiol 2: 641–646.
Poly F, Monrozier LJ, Bally R . (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152: 95–103.
Ramírez-Saad H, Akkermans WL, Akkermans DL . (1996). DNA extraction from actinorhizal nodules. In: Akkermans, ADL, van Elsas JD, de Bruijn, FJ (eds). Molecular Microbial Ecology Manual. Kluver Academic Publishers: Dordrecht. pp 1–11.
Rigaud J, Puppo A . (1975). Indole-3-acetic catabolism by soybean bacteroids. J Gen Microbiol 8: 223–228.
Rivas R, Martens M, Ladujie P, Willems A . (2009). Multilocus sequence analysis of the genus Bradyrhizobium. System Appl Microbiol 32: 101–110.
Rivas R, Sanchez M, Trujillo ME, Zurdo-Pineiro JL, Mateos PF, Martinez-Molina E et al. (2003). Xylanimonas cellulosilytica gen. nov., sp. nov., a xylanolytic bacterium isolated from a decayed tree (Ulmus nigra). Int J Syst Evol Microbiol 53: 99–103.
Rodríguez R . (2008). Análisis de la población bacteriana endofita presente en nódulos de Lupinus: interacción y localización in situ. Ph.D. Thesis. Universidad de Salamanca, Spain.
Rosenblueth M, Martinez-Romero E . (2006). Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19: 827–837.
Saitou N, Nei M . (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425.
Shirling EB, Gottlieb D . (1966). Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16: 313–340.
Stepkowski T, Hughes CE, Law IJ, Markiewicz L, Gurda D, Chlebicka A et al. (2007). Diversification of lupine Bradyrhizobium strains: evidence from nodulation gene trees. Appl Environ Microbiol 73: 3254–3264.
Tamura K, Dudley J, Nei M, Kumar S . (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
Thawai C, Kittakoop P, Tanasupawat S, Suwanborirux K, Sriklung K, Thebtaranonth Y . (2004). Micromonosporin A, a novel 24-membered polyene lactam macrolide from Micromonospora sp. isolated from peat swamp forest. Chem Biodivers 1: 640–645.
Thawai C, Tanasupawat S, Itoh T, Suwanborirux K, Kudo T . (2005). Micromonospora siamensis sp. nov., isolated from Thai peat swamp forest. J Gen Appl Microbiol 51: 229–234.
Thawai C, Tanasupawat S, Suwanborirux K, Itoh T, Kudo T . (2007). Micromonospora narathiwatensis sp. nov., from Thai peat swamp forest soils. J Gen Appl Microbiol 53: 287–293.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG . (1997). The clustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res 24: 4876–4882.
Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA et al. (2002). Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68: 2161–2171.
Torres AR, Araujo WL, Cursino L, Hungria M, Plotegher F, Mostasso FL et al. (2008). Diversity of endophytic enterobacteria associated with different host plants. J Microbiol 46: 373–379.
Trujillo ME, Kroppenstedt RM, Fernandez-Molinero C, Schumann P, Martinez-Molina E . (2007). Micromonospora lupini sp. nov. and Micromonospora saelicesensis sp. nov., isolated from root nodules of Lupinus angustifolius. Int J Syst Evol Microbiol 57: 2799–2804.
Trujillo ME, Kroppenstedt RM, Schumann P, Carro L, Martinez-Molina E . (2006). Micromonospora coriariae sp. nov., isolated from root nodules of Coriaria myrtifolia. Int J Syst Evol Microbiol 56: 2381–2385.
Valdés M, Perez NO, Estrada-de Los Santos P, Caballero-Mellado J, Pena-Cabriales JJ, Normand P et al. (2005). Non-Frankia actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl Environ Microbiol 71: 460–466.
Versalovic J, Scheider M, De Bruijn FJ, Lupski JR . (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Meth Mol Cell Biol 5: 25–40.
Vincent JM . (1970). The cultivation, isolation and maintenance of rhizobia. In: Vincent JM (eds). A Manual for the Practical Study of Root Nodule Bacteria. Blackwell Scientific: Oxford. pp 1–13.
Vinuesa P, Leon-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Perez-Galdona R et al. (2005). Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol 55: 569–575.
Walkley A . (1947). A critical examination of rapid method for determining organic carbon soil-effect of variations in digestión condition and inorganic soil constituents. Soil Sci 63: 251–264.
Zakhia F, Jeder H, Willems A, Gillis M, Dreyfus B, de Lajudie P . (2006). Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb Ecol 51: 375–393.
Zhao H, Kassama Y, Young M, Kell DB, Goodacre R . (2004). Differentiation of Micromonospora isolates from a coastal sediment in Wales on the basis of Fourier transform infrared spectroscopy, 16S rRNA sequence analysis, and the amplified fragment length polymorphism technique. Appl Environ Microbiol 70: 6619–6627.
Acknowledgements
This work was funded by the Ministerio de Educación y Ciencia under the project CGL2006-06988 to MET and EMM. Ph.D. grants are also acknowledged to the following supporting bodies: Ministerio de Educación y Ciencia (PAV, AP20052608), Universidad de Pamplona, Colombia (RR), Junta de Castilla y León (LC), and Banco Santander (EC). We thank Carmelo Avila for his advice in statistical analyses; K. Findlay and M. Ortiz for carrying out microscopy work and Ann M. Hirsch for her valuable comments. We thank three anonymous reviewers for their constructive comments and recommendations.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Trujillo, M., Alonso-Vega, P., Rodríguez, R. et al. The genus Micromonospora is widespread in legume root nodules: the example of Lupinus angustifolius. ISME J 4, 1265–1281 (2010). https://doi.org/10.1038/ismej.2010.55
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.55
Keywords
This article is cited by
-
Endophytic actinobacteria from wild medicinal plants are a natural source of insecticide to control the African cotton leafworm (Spodoptera littoralis)
AMB Express (2023)
-
Effect of Micromonospora sp. KSC08 on nitrogen conservation throughout composting
Biomass Conversion and Biorefinery (2023)
-
Bioactivities of endophytic actinobacteria inhabiting Artemisia herba-alba emphasizing differences from free-living strains
Folia Microbiologica (2022)
-
Medicago root nodule microbiomes: insights into a complex ecosystem with potential candidates for plant growth promotion
Plant and Soil (2022)
-
Isolation and characterisation of endophytic actinobacteria and their effect on the growth and nodulation of chickpea (Cicer arietinum)
Plant and Soil (2021)