Abstract
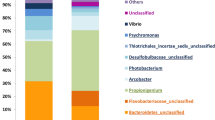
Microorganisms are globally dispersed and are able to proliferate in any habitat that supports their lifestyles, which, however, has not yet been explored in any specific microbial taxon. The social myxobacteria are considered typical soil bacteria because they have been identified in various terrestrial samples, a few in coastal areas, but none in other oceanic environments. To explore the prevalence of marine myxobacteria and to investigate their phylogenetic relationships with their terrestrial counterparts, we established myxobacteria-enriched libraries of 16S rRNA gene sequences from four deep-sea sediments collected at depths from 853 to 4675 m and a hydrothermal vent at a depth of 204 m. In all, 68 different myxobacteria-related sequences were identified from randomly sequenced clones of the libraries of different samples. These myxobacterial sequences were diverse but phylogenetically similar at different locations and depths. However, they were separated from terrestrial myxobacteria at high levels of classification. This discovery indicates that the marine myxobacteria are phylogeographically separated from their terrestrial relatives, likely because of geographic separation and environment selection.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Andersson AF, Riemann L, Bertilsson S . (2010). Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J 4: 171–181.
Arakawa S, Sato T, Sato R, Zhang J, Gamo T, Tsunogai U et al. (2006). Molecular phylogenetic and chemical analyses of the microbial mats in deep-sea cold seep sediments at the northeastern Japan Sea. Extremophiles 10: 311–319.
Bowman JP, Rea SM, McCammon SA, McMeekin TA . (2000). Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hilds, Eastern Antarctica. Environ Microbiol 2: 227–237.
Dawid W . (2000). Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev 24: 403–427.
Dworkin M, Kaiser D . (1993). Myxobacteria II. ASM Press: Washington, DC.
Fudou R, Jojima Y, Iizuka T, Yamanaka S . (2002). Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. J Gen Appl Microbiol 48: 109–116.
Fuhrman JA . (2009). Microbial community structure and its functional implications. Nature 459: 193–199.
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C . (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA 106: 22427–22432.
Gerth K, Pradella S, Perlova O, Beyer S, Müller R . (2003). Myxobacteria: proficient producers of novel natural products with various biological activities—past and future biotechnological aspects with the focus on the genus Sorangium. J Biotechnol 106: 233–253.
Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Green JL, Bohannan BJM, Whitaker RJ . (2008). Microbial biogeography: from taxonomy to traits. Science 320: 1039–1043.
Iizuka T, Jojima Y, Fudou R, Hiraishi A, Ahn JW, Yamanaka S . (2003a). Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int J Syst Evol Microbiol 53: 189–195.
Iizuka T, Jojima Y, Fudou R, Tokura M, Hiraishi A, Yamanaka S . (2003b). Enhygromyxa salina gen. nov., sp. nov., a slightly halophilic myxobacterium isolated from the coastal areas of Japan. Syst Appl Microbiol 26: 189–196.
Iizuka T, Jojima Y, Fudou R, Yamanaka S . (1998). Isolation of myxobacteria from the marine environment. FEMS Microbiol Lett 169: 317–322.
Jiang DM, Wu ZH, Zhao JY, Li YZ . (2007). Fruiting and non-fruiting myxobacteria: a phylogenetic perspective of cultured and uncultured members of this group. Mol Phylogenet Evol 44: 545–552.
Kumar S, Tamura K, Nei M . (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings Bioinformat 5: 150–163.
Li YZ, Hu W, Zhang YQ, Qiu ZJ, Zhang Y, Wu BH . (2002). A simple method to isolate salt-tolerant myxobacteria from marine samples. J Microbiol Methods 50: 205–209.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440.
Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC et al. (2005). Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol 55: 1759–1766.
Martiny JB, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
Moyer CL, Dobbs FC, Karl DM . (1995). Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol 61: 1555–1562.
Pan HW, Liu H, Liu T, Li CY, Li ZF, Cai K et al. (2009). Seawater-regulated genes for two-component systems and outer membrane proteins in Myxococcus. J Bacteriol 191: 2102–2111.
Pham VD, Konstantinidis KT, Palden T, DeLong EF . (2008). Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol 10: 2313–2330.
Ravenschlag K, Sahm K, Pernthaler J, Amann R . (1999). High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65: 3982–3989.
Reichenbach H . (1999). The ecology of the myxobacteria. Environ Microbiol 1: 15–21.
Reichenbach H . (2001). Myxobacteria, producers of novel bioactive substances. J Ind Microbiol Biotechnol 27: 149–156.
Reichenbach H . (2004). The Myxococcales. Bergey's Manual of Systematic Bacteriology, Garrity GM (ed). Springer-Verlag: New York, NY, pp 1059–1143.
Reichenbach H, Dworkin M . (1992). The myxobacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds). The Prokaryotes. 2nd ed. Springer-Verlag: New York, NY, pp 3416–3487.
Rosenberg E . (1984). Myxobacteria: Development and Cell Interactions. Springer-Verlag: New York, NY.
Sanford RA, Cole JR, Tiedje JM . (2002). Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol 68: 893–900.
Shimkets LJ . (1990). Social and developmental biology of myxobacteria. Microbiol Rev 54: 473–501.
Shimkets LJ, Dworkin M, Reichenbach H . (2005). The myxobacteria. The Prokaryotes 3rd edn (release 3.19) Springer-Verlag: New York, NY.
Spröer C, Reichenbach H, Stackebrandt E . (1999). The correlation between morphogenetic classification of myxobacteria. Int J Syst Bacteriol 49: 1255–1262.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG . (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882.
von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ et al. (2007). Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315: 1126–1130.
Wang B, Hu W, Liu H, Zhang CY, Zhao JY, Jiang DM et al. (2007a). Adaptation of salt-tolerant Myxococcus strains and their motility systems to the ocean conditions. Microb Ecol 54: 43–51.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007b). Naive Bayesian Classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Whitman WB, Coleman DC, Wiebe WJ . (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583.
Whitworth DE . (2007). Myxobacteria: Multicellularity and Differentiation. ASM Press: Washington, DC.
Wu ZH, Jiang DM, Li P, Li YZ . (2005). Exploring the diversity of myxobacteria in a soil niche by myxobacteria-specific primers and probes. Environ Microbiol 7: 1602–1610.
Zhang CY, Cai K, Liu H, Zhang Y, Pan HW, Wang B et al. (2007). A new locus important for Myxococcus social motility and development. J Bacteriol 189: 7937–7941.
Zhang J, Liu Z, Wang S, Jiang P . (2002). Characterization of a bioflocculant produced by the marine myxobacterium Nannocystis sp. NU-2. Appl Microbiol Biotechnol 59: 517–522.
Zhang W, Ki J-S, Qian P-Y . (2008). Microbial diversity in polluted harbor sediments I: bacterial community assessment based on four clone libraries of 16S rDNA. Estuar Coast Shelf Sci 76: 668–681.
Zhang YQ, Li YZ, Wang B, Wu ZZ, Zhang CZ, Gong X et al. (2005). Characteristics and living patterns of marine myxobacterial isolates. Appl Environ Microbiol 71: 3331–3336.
Acknowledgements
This work was financially supported by National Science Foundation for Distinguished Young Scholars (No. 30825001), National Natural Science Foundation (No. 30671192 and 30870001) and National High Technology Research and Development program (No. 2007AA021501) of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Jiang, DM., Kato, C., Zhou, XW. et al. Phylogeographic separation of marine and soil myxobacteria at high levels of classification. ISME J 4, 1520–1530 (2010). https://doi.org/10.1038/ismej.2010.84
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.84
Keywords
This article is cited by
-
Bradymonabacteria, a novel bacterial predator group with versatile survival strategies in saline environments
Microbiome (2020)
-
Analysis of the Genome and Metabolome of Marine Myxobacteria Reveals High Potential for Biosynthesis of Novel Specialized Metabolites
Scientific Reports (2018)
-
Current trends in myxobacteria research
Annals of Microbiology (2016)
-
Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone
The ISME Journal (2014)
-
Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu
Scientific Reports (2013)