Abstract
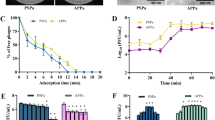
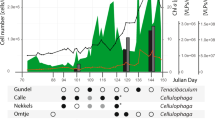
Marine phages have an astounding global abundance and ecological impact. However, little knowledge is derived from phage genomes, as most of the open reading frames in their small genomes are unknown, novel proteins. To infer potential functional and ecological relevance of sequenced marine Pseudoalteromonas phage H105/1, two strategies were used. First, similarity searches were extended to include six viral and bacterial metagenomes paired with their respective environmental contextual data. This approach revealed ‘ecogenomic’ patterns of Pseudoalteromonas phage H105/1, such as its estuarine origin. Second, intrinsic genome signatures (phylogenetic, codon adaptation and tetranucleotide (tetra) frequencies) were evaluated on a resolved intra-genomic level to shed light on the evolution of phage functional modules. On the basis of differential codon adaptation of Phage H105/1 proteins to the sequenced Pseudoalteromonas spp., regions of the phage genome with the most ‘host’-adapted proteins also have the strongest bacterial tetra signature, whereas the least ‘host’-adapted proteins have the strongest phage tetra signature. Such a pattern may reflect the evolutionary history of the respective phage proteins and functional modules. Finally, analysis of the structural proteome identified seven proteins that make up the mature virion, four of which were previously unknown. This integrated approach combines both novel and classical strategies and serves as a model to elucidate ecological inferences and evolutionary relationships from phage genomes that typically abound with unknown gene content.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Abuladze NK, Gingery M, Tsai J, Eiserling FA . (1994). Tail length determination in bacteriophage T4. Virology 199: 301–310.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Andersson AF, Banfield JF . (2008). Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050.
Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C et al. (2006). The marine viromes of four oceanic regions. PLoS Biol 4: e368.
Bahir I, Fromer M, Prat Y, Linial M . (2009). Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol Syst Biol 5: 311.
Becker GAB, Dick SD, Dippner JWD . (1992). Hydrography of the German Bight. Mar Ecol Prog Ser 91: 9–19.
Besemer J, Lomsadze A, Borodovsky M . (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29: 2607–2618.
Black LW . (1989). DNA Packaging in dsDNA Bacteriophages. Annu Rev Microbiol 43: 267–292.
Borbély G, Kaki C, Gulyás A, Farkas GL . (1980). Bacteriophage infection interferes with guanosine 3′-diphosphate-5′-diphosphate accumulation induced by energy and nitrogen starvation in the cyanobacterium Anacystis nidulans. J Bacteriol 144: 859–864.
Botstein D . (1980). A theory of modular evolution for bacteriophages. Ann NY Acad Sci 354: 484–491.
Botstein D, Matz MJ . (1970). A recombination function essential to the growth of bacteriophage P22. J Mol Biol 54: 417–440.
Broad Institute (2010). Marine Phage Sequencing Project##http://www.broadinstitute.org/annotation/viral/Phage.
Bryan MJ, Burroughs NJ, Spence EM, Clokie MR, Mann NH, Bryan SJ . (2008). Evidence for the intense exchange of MazG in marine cyanophages by horizontal gene transfer. PLoS One 3: e2048.
Bushman F . (2002). Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY.
Calendar R . (1970). The regulation of phage development. Annu Rev Microbiol 24: 241–296.
Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H . (2003). Prophage genomics. Microbiol Mol Biol R 67: 238.
Carbone A . (2008). Codon bias is a major factor explaining phage evolution in translationally biased hosts. J Mol Evol 66: 210–223.
Carbone A, Zinovyev A, Képès F . (2003). Codon adaptation index as a measure of dominating codon bias. Bioinformatics 19: 2005–2015.
Casjens S . (2003). Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49: 277–300.
Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML . et al. (2005). The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J Bacteriol 187: 1091–1104.
Clokie MR, Mann NH . (2006). Marine cyanophages and light. Environ Microbiol 8: 2074–2082.
Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85.
Duffy S, Turner PE . (2008). Phage evolutionary biology. In: Abedon, S. (ed). Bacteriophage Ecology: Population Growth, Evolution, and Impact of Bacterial Viruses. Cambridge University Press: Cambridge, UK, pp 147–176.
Emanuelsson O, Brunak S, von Heijne G, Nielsen H . (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971.
Erskine JM . (1973). Characteristics of Erwinia amylovora bacteriophage and its possible role in the epidemology of fire blight. Can J Microbiol 19: 837–845.
Field D . (2008). Working together to put molecules on the map. Nature 453: 978.
Filée J, Bapteste E, Susko E, Krisch HM . (2006). A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Mol Biol Evol 23: 1688–1696.
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE et al. (2010). The Pfam protein families database. Nucleic Acids Res 38: D211–D222.
Friga GM, Borbély G, Farkas GL . (1981). Accumulation of guanosine tetraphosphate (ppGpp) under nitrogen starvation in Anacystis nidulans, a cyanobacterium. Arch Microbiol 129: 341–343.
Gross M, Marianovsky I, Glaser G . (2006). MazG—a regulator of programmed cell death in Escherichia coli. Mol Microbiol 59: 590–601.
Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC et al. (2005). JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33: W526–W531.
Haggard-Ljungquist E, Halling C, Calendar R . (1992). DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol 174: 1462.
Hendrix RW, Smith M, Burns RN, Ford ME, Hatfull GF . (1999). Evolutionary relationships among diverse bacteriophages and prophages: all the world’sa phage. PNAS 96: 2192.
Iyer LM, Koonin EV, Aravind L . (2002). Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3: 8.
Kottmann R, Kostadinov I, Duhaime MB, Buttigieg PL, Yilmaz P, Hankeln W et al. (2010). Megx.net: integrated database resource for marine ecological genomics. Nucleic Acids Res 38: D391–D395.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL . (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948.
Lima-Mendez G, Van Helden J, Toussaint A, Leplae R . (2008). Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24: 863–865.
Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW . (2005). Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438: 86–89.
Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW . (2004). Transfer of photosynthesis genes to and from Prochlorococcus viruses. PNAS 101: 11013.
Long A, McDaniel LD, Mobberley J, Paul JH . (2008). Comparison of lysogeny (prophage induction) in heterotrophic bacterial and Synechococcus populations in the Gulf of Mexico and Mississippi River plume. ISME J 2: 132–144.
Lucchini S, Desiere F, Brüssow H . (1999). Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J Virol 73: 8647–8656.
Lucks JB, Nelson DR, Kudla GR, Plotkin JB . (2008). Genome landscapes and bacteriophage codon usage. PLoS Comput Biol 4: e1000001.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Männistö RH, Kivelä HM, Paulin L, Bamford DH, Bamford JK . (1999). The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262: 355–363.
McDaniel L, Breitbart M, Mobberley J, Long A, Haynes M, Rohwer F et al. (2008). Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS One 3: e3263.
Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN et al. (2005). Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res 15: 1325–1335.
Miller RV, Day MJ . (2008). Contribution of lysogeny, pseudolysogeny, and starvation to phage ecology. In: Abedon, S. (ed). Bacteriophage Ecology: Population Growth, Evolution, and Impact of Bacterial Viruses. Cambridge University Press: Cambridge, UK, pp 114–143.
Mobberley JM, Authement RN, Segall AM, Paul JH . (2008). The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J Virol 82: 6618–6630.
Moebus K . (1992). Further investigations on the concentration of marine bacteriophages in the water around Helgoland, with reference to the phage-host systems encountered. Helgoland Mar Res 46: 275–292.
Moebus K . (1997). Investigations of the marine lysogenic bacterium H24. 2. Development of pseudolysogeny in nutrient rich broth. Mar Ecol Prog Ser 148: 229–240.
Muyzer G, Teske A, Wirsen CO, Jannasch HW . (1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164: 165–172.
Paul JH, Sullivan MB, Segal AM, Rohwer F . (2002). Marine phage genomics. Comp Biochem Phys 133: 463–476.
Paul JH, Williamson SJ, Long A, Authement RN, John D, Segall AM et al. (2005). Complete genome sequence of phiHSIC, a pseudotemperate marine phage of Listonella pelagia. Appl Environ Microbiol 71: 3311–3320.
Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA et al. (2003). Origins of highly mosaic mycobacteriophage genomes. Cell 113: 171–182.
Poteete AR, Sauer RT, Hendrix RW . (1983). Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J Mol Biol 171: 401–418.
Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ . (2003). Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res 13: 145–158.
Pride DT, Wassenaar TM, Ghose C, Blaser MJ . (2006). Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics 7: 8.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Ptashne M . (2004). A Genetic Switch. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY.
Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW . (2000). Genomic sequence and analysis of the atypical temperate bacteriophage N15. J Mol Biol 299: 53–73.
Richter M, Lombardot T, Kostadinov I, Kottmann R, Duhaime MB, Peplies J et al. (2008). Jcoast—a biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes. BMC Bioinformatics 9: 177.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77.
Sambrook J, Russell DW . (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York.
Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M . (2007). CAMERA: a community resource for metagenomics. PLoS Biol 5: e75.
Sharp PM, Li WH . (1987). The Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15: 1281–1295.
Siefert JL . (2009). Defining the mobilome. Methods Mol Biol 532: 13–27.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW . (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144.
Suttle CA . (2007). Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5: 801–812.
Teeling H, Meyerdierks A, Bauer M, Amann R, Glöckner FO . (2004). Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol 6: 938–947.
Thomas T, Evans FF, Schleheck D, Mai-Prochnow A, Burke C, Penesyan A et al. (2008). Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS One 3: e3252.
Toussaint A, Lima-Mendez G, Leplae R . (2007). PhiGO, a phage ontology associated with the ACLAME database. Res Microbiol 158: 567–571.
Vandenbergh PA, Cole RL . (1986). Cloning and Expression in Escherichia coli of the Polysaccharide Depolymerase Associated with Bacteriophage-Infected Erwinia amylovora. Appl Environ Microbiol 51: 862–864.
Waldmann J . (2010). ocount2: a library for the calculation and comparison of oligonucleotide patterns##http://www.promedici.de/ocount2.
Wang IN, Smith DL, Young R . (2000). Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54: 799–825.
Wichels A, Biel SS, Gelderblom HR, Brinkhoff T, Muyzer G, Schütt C . (1998). Bacteriophage diversity in the North Sea. Appl Environ Microbiol 64: 4128–4133.
Wichels A, Gerdts G, Schütt C . (2002). Pseudoalteromonas spp. phages, a significant group of marine bacteriophages in the North Sea. Aquat Microb Ecol 27: 233–239.
Williamson SJ, McLaughlin MR, Paul JH . (2001). Interaction of the PhiHSIC virus with its host: lysogeny or pseudolysogeny? Appl Environ Microbiol 67: 1682–1688.
Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO . et al. (2006). Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443: 950–955.
Zhong Y, Chen F, Wilhelm SW, Poorvin L, Hodson RE . (2002). Phylogenetic Diversity of Marine Cyanophage Isolates and Natural Virus Communities as Revealed by Sequences of Viral Capsid Assembly Protein Gene gp20. AEM 68: 4.
Acknowledgements
MBD is grateful to Amelia Rotaru for SDS-PAGE assistance, Marianne Jacob for contributions to tetra analysis, Matthew B Sullivan and Vincent Denef for critically reading the paper, and Matthias Ullrich, Renzo Kottmann and Ivaylo Kostadinov for fruitful discussions and support. A Marie Curie Early Stage Training Fellowship to MBD supports funding for this project (MEST-CT-2004-007776) through the MarMic program of the MPI for Marine Microbiology. Further acknowledgement goes to two anonymous reviewers who offered useful suggestions, strengthening this analysis and paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Duhaime, M., Wichels, A., Waldmann, J. et al. Ecogenomics and genome landscapes of marine Pseudoalteromonas phage H105/1. ISME J 5, 107–121 (2011). https://doi.org/10.1038/ismej.2010.94
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.94
Keywords
This article is cited by
-
Isolation and Complete Sequence of One Novel Marine Bacteriophage PHS21 Infecting Pseudoalteromonas marina
Current Microbiology (2022)
-
Phage-specific metabolic reprogramming of virocells
The ISME Journal (2020)
-
Isolation and Genome Sequencing of a Novel Pseudomonas aeruginosa Phage PA-YS35
Current Microbiology (2020)
-
Isolation and Complete Genome of the Marine Pseudoalteromonas Phage C7 from Coastal Seawater of Yellow Sea, China
Current Microbiology (2020)
-
Host-hijacking and planktonic piracy: how phages command the microbial high seas
Virology Journal (2019)