Abstract
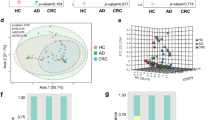
Despite a long-suspected role in the development of human colorectal cancer (CRC), the composition of gut microbiota in CRC patients has not been adequately described. In this study, fecal bacterial diversity in CRC patients (n=46) and healthy volunteers (n=56) were profiled by 454 pyrosequencing of the V3 region of the 16S ribosomal RNA gene. Both principal component analysis and UniFrac analysis showed structural segregation between the two populations. Forty-eight operational taxonomic units (OTUs) were identified by redundancy analysis as key variables significantly associated with the structural difference. One OTU closely related to Bacteroides fragilis was enriched in the gut microbiota of CRC patients, whereas three OTUs related to Bacteroides vulgatus and Bacteroides uniformis were enriched in that of healthy volunteers. A total of 11 OTUs belonging to the genera Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus and Peptostreptococcus were significantly more abundant in the gut microbiota of CRC patients, and 5 OTUs belonging to the genus Roseburia and other butyrate-producing bacteria of the family Lachnospiraceae were less abundant. Real-time quantitative PCR further validated the significant reduction of butyrate-producing bacteria in the gut microbiota of CRC patients by measuring the copy numbers of butyryl-coenzyme A CoA transferase genes (Mann–Whitney test, P<0.01). Reduction of butyrate producers and increase of opportunistic pathogens may constitute a major structural imbalance of gut microbiota in CRC patients.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L . (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3: e2836.
Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS . (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol 23 (Part 1): 1298–1303.
Bernstein H, Bernstein C, Payne CM, Dvorak K . (2009). Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol 15: 3329–3340.
Biarc J, Nguyen IS, Pini A, Gosse F, Richert S, Thierse D et al. (2004). Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis 25: 1477–1484.
Bingham SA . (2000). Diet and colorectal cancer prevention. Biochem Soc Trans 28: 12–16.
Borody TJ, Warren EF, Leis S, Surace R, Ashman O . (2003). Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol 37: 42–47.
Chan AT, Giovannucci EL . (2010). Primary prevention of colorectal cancer. Gastroenterology 138: 2029–2043, e2010.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37 (Database issue): D141–D145.
Collins D, Hogan AM, Winter DC . (2010). Microbial and viral pathogens in colorectal cancer. Lancet Oncol 12: 504–512.
Dahm CC, Keogh RH, Spencer EA, Greenwood DC, Key TJ, Fentiman IS et al. (2010). Dietary fiber and colorectal cancer risk: a nested case-control study using food diaries. J Natl Cancer Inst 102: 614–626.
Davis CD, Milner JA . (2009). Gastrointestinal microflora, food components and colon cancer prevention. J Nutr Biochem 20: 743–752.
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107: 14691–14696.
DeSantis Jr TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al. (2006). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34 (Web Server issue): W394–W399.
Dethlefsen L, Huse S, Sogin ML, Relman DA . (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol 6: e280.
Devillard E, McIntosh FM, Duncan SH, Wallace RJ . (2007). Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol 189: 2566–2570.
Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE . (2007). Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73: 1073–1078.
Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP et al. (2000). Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis 21: 753–756.
Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR . (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785.
Good IJ . (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264.
Hamady M, Lozupone C, Knight R . (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME 4: 17–27.
Homann N, Tillonen J, Salaspuro M . (2000). Microbially produced acetaldehyde from ethanol may increase the risk of colon cancer via folate deficiency. Int J Cancer 86: 169–173.
Huycke MM, Abrams V, Moore DR . (2002). Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23: 529–536.
Huycke MM, Gaskins HR . (2004). Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med (Maywood) 229: 586–597.
Huycke MM, Moore DR . (2002). In vivo production of hydroxyl radical by Enterococcus faecalis colonizing the intestinal tract using aromatic hydroxylation. Free Radic Biol Med 33: 818–826.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . (2011). Global cancer statistics. CA Cancer J Clin 61: 69–90.
Kalliomaki M, Isolauri E . (2003). Role of intestinal flora in the development of allergy. Curr Opin Allergy Clin Immunol 3: 15–20.
Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H et al. (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA 105: 2117–2122.
Li W, Godzik A . (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659.
Louis P, Flint HJ . (2007). Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol 73: 2009–2012.
Louis P, Flint HJ . (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294: 1–8.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Lynch HT, de la Chapelle A . (2003). Hereditary colorectal cancer. N Engl J Med 348: 919–932.
Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ . (2009). Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One 4: e5517.
Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R et al. (2004). Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127: 80–93.
Mazmanian SK, Round JL, Kasper DL . (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453: 620–625.
Moore WE, Moore LH . (1995). Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol 61: 3202–3207.
Muyzer G, de Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700.
Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y . (2007). Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol 56 (Part 12): 1669–1674.
Nicholson JK, Holmes E, Wilson ID . (2005). Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol 3: 431–438.
O’Keefe SJD, Chung D, Mahmoud N, Sepulveda AR, Manafe M, Arch J et al. (2007). Why do African Americans get more colon cancer than Native Africans. J Nutr 137: 175S–182S.
Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR et al. (2004). Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53: 685–693.
Palombo JD, Ganguly A, Bistrian BR, Menard MP . (2002). The antiproliferative effects of biologically active isomers of conjugated linoleic acid on human colorectal and prostatic cancer cells. Cancer Lett 177: 163–172.
Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ . (2002). The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217: 133–139.
Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P . (2009). Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101: 541–550.
Rose DJ, DeMeo MT, Keshavarzian A, Hamaker BR . (2007). Influence of dietary fiber on inflammatory bowel disease and colon cancer: importance of fermentation pattern. Nutr Rev 65: 51–62.
Saleh M, Trinchieri G . (2011). Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol 11: 9–20.
Scanlan PD, Shanahan F, Marchesi JR . (2009). Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol 69: 213–221.
Scharlau D, Borowicki A, Habermann N, Hofmann T, Klenow S, Miene C et al. (2009). Mechanisms of primary cancer prevention by butyrate and other products formed during gut flora-mediated fermentation of dietary fibre. Mutat Res 682: 39–53.
Schloss PD, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N et al. (2010). Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 1: 138–147.
Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P et al. (2011). Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6: e16393.
Terzic J, Grivennikov S, Karin E, Karin M . (2010). Inflammation and colon cancer. Gastroenterology 138: 2101–2114, e2105.
Tong JL, Ran ZH, Shen J, Fan GQ, Xiao SD . (2008). Association between fecal bile acids and colorectal cancer: a meta-analysis of observational studies. Yonsei Med J 49: 792–803.
Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C . (2009). Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One 4: e6026.
van Hylckama Vlieg JE, Veiga P, Zhang C, Derrien M, Zhao L . (2011). Impact of microbial transformation of food on health-from fermented foods to fermentation in the gastro-intestinal tract. Curr Opin Biotechnol 22: 211–219.
Waidmann M, Bechtold O, Frick JS, Lehr HA, Schubert S, Dobrindt U et al. (2003). Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125: 162–177.
Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X et al. (2010). Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5: 220–230.
Wei H, Dong L, Wang T, Zhang M, Hua W, Zhang C et al. (2010). Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol Ecol 73: 577–586.
Willing B, Dicksved J, Halfvarson J, Andersson A, Lucio M, Zheng Z et al. (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139: 1844–1854, e1.
Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L et al. (2009). Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis 15: 653–660.
Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17T cell responses. Nat Med 15: 1016–1022.
Zhang CH, Zhang MH, Wang SY, Han RJ, Cao YF, Hua WY et al. (2010). Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 4: 232–241.
Zhang ML, Zhang MH, Zhang CH, Du HM, Wei GF, Pang XY et al. (2009). Pattern extraction of structural responses of gut microbiota to rotavirus infection via multivariate statistical analysis of clone library data. FEMS Microbiol Ecol 70: 177–185.
Acknowledgements
This work was supported by Project 2006BAI11B08-02 of the National Science & Technology Pillar Program, Key Project 30730005 of the National Nature Science Foundation of China (NSFC), Project 2009ZX10004-601 of National Science and Technology Major Project of China and Key Project 2007CB513002 of Chinese National Programs for Fundamental Research and Development (973 program).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Wang, T., Cai, G., Qiu, Y. et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6, 320–329 (2012). https://doi.org/10.1038/ismej.2011.109
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.109
Keywords
This article is cited by
-
Analysis of differences in intestinal flora associated with different BMI status in colorectal cancer patients
Journal of Translational Medicine (2024)
-
Endogenous Coriobacteriaceae enriched by a high-fat diet promotes colorectal tumorigenesis through the CPT1A-ERK axis
npj Biofilms and Microbiomes (2024)
-
Effect of periodontitis induced by Fusobacterium nucleatum on the microbiota of the gut and surrounding organs
Odontology (2024)
-
Modulation of intestinal metabolites by calorie restriction and its association with gut microbiota in a xenograft model of colorectal cancer
Discover Oncology (2024)
-
Percutaneous transhepatic cholangial drainage or antibiotic therapy worsens response to immunotherapy in advanced cholangiocarcinoma
BMC Cancer (2023)