Abstract
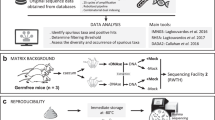
To determine the reproducibility and quantitation of the amplicon sequencing-based detection approach for analyzing microbial community structure, a total of 24 microbial communities from a long-term global change experimental site were examined. Genomic DNA obtained from each community was used to amplify 16S rRNA genes with two or three barcode tags as technical replicates in the presence of a small quantity (0.1% wt/wt) of genomic DNA from Shewanella oneidensis MR-1 as the control. The technical reproducibility of the amplicon sequencing-based detection approach is quite low, with an average operational taxonomic unit (OTU) overlap of 17.2%±2.3% between two technical replicates, and 8.2%±2.3% among three technical replicates, which is most likely due to problems associated with random sampling processes. Such variations in technical replicates could have substantial effects on estimating β-diversity but less on α-diversity. A high variation was also observed in the control across different samples (for example, 66.7-fold for the forward primer), suggesting that the amplicon sequencing-based detection approach could not be quantitative. In addition, various strategies were examined to improve the comparability of amplicon sequencing data, such as increasing biological replicates, and removing singleton sequences and less-representative OTUs across biological replicates. Finally, as expected, various statistical analyses with preprocessed experimental data revealed clear differences in the composition and structure of microbial communities between warming and non-warming, or between clipping and non-clipping. Taken together, these results suggest that amplicon sequencing-based detection is useful in analyzing microbial community structure even though it is not reproducible and quantitative. However, great caution should be taken in experimental design and data interpretation when the amplicon sequencing-based detection approach is used for quantitative analysis of the β-diversity of microbial communities.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Acosta-Martinez V, Dowd S, Sun Y, Allen V . (2008). Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Biochem 40: 2762–2770.
Ahn SJ, Costa J, Emanuel JR . (1996). PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res 24: 2623–2625.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R et al. (2007). The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PloS ONE 2: e197.
Chao A . (1987). Estimating the population-size for capture recapture data with unequal catchability. Biometrics 43: 783–791.
Cheung MK, Au CH, Chu KH, Kwan HS, Wong CK . (2010). Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J 4: 1053–1059.
Chun J, Kim KY, Lee JH, Choi Y . (2010). The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol 10: 101.
Clarke KR . (1993). Nonparametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143.
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM et al. (2007). The Ribosomal Database Project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Daniel R . (2005). The metagenomics of soil. Nat Rev Microbiol 3: 470–478.
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM et al. (2006). Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7: 57.
Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H et al. (2010). Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4: 642–647.
Gomez-Alvarez V, Teal TK, Schmidt TM . (2009). Systematic artifacts in metagenomes from complex microbial communities. ISME J 3: 1314–1317.
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R . (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237.
He Z, Deng Y, Van Nostrand JD, Tu QC, Xu MY, Hemme CL et al. (2010a). GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4: 1167–1179.
He Z, Gentry TJ, Schadt CW, Wu LY, Liebich J, Chong SC et al. (2007). GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1: 67–77.
He Z, Xu MY, Deng Y, Kang SH, Kellogg L, Wu LY et al. (2010b). Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett 13: 564–575.
He ZL, Van Nostrand JD, Wu LY, Zhou JZ . (2008). Development and application of functional gene arrays for microbial community analysis. Transactions of Nonferrous Metals Society of China 18: 1319–1327.
Hill MO, Gauch HG . (1980). Detrended correspondence-analysis – an improved ordination technique. Vegetation 42: 47–58.
Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ . (2010). Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4: 829–838.
Huber JA, Mark Welch D, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D . (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143.
Huse SM, Welch DM, Morrison HG, Sogin ML . (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898.
Iwai S, Chai BL, Sul WJ, Cole JR, Hashsham SA, Tiedje JM . (2010). Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J 4: 279–285.
Koopman MM, Fuselier DM, Hird S, Carstens BC . (2010). The carnivorous pale pitcher plant harbors diverse, distinct, and time-dependent bacterial communities. Appl Environ Microbiol 76: 1851–1860.
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P . (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Luo YQ, Wan SQ, Hui DF, Wallace LL . (2001). Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413: 622–625.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
McCune B, Grace JB . (2002). Analysis of Ecological Communities. MJM Software Design: Gleneden Beach, OR.
McCune B, Mefford MJ . (1999). PC-ORD: Multivariate Analysis of Ecological Data. Version 4. User's guide MJM Software Design: Gleneden Beach, OR.
Mielke PW, Berry KJ . (2001). Permutation Methods: A Distance Function Approach. Springer Series in Statistics. Springer: New York, NY.
Nawrocki EP, Eddy SR . (2007). Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput Biol 3: 540–554.
Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M et al. (2007). A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35: e130.
Qiu XY, Wu LY, Huang HS, McDonel PE, Palumbo AV, Tiedje JM et al. (2001). Evaluation of PCR-generated chimeras: mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl Environ Microbiol 67: 880–887.
Quince C, Curtis TP, Sloan WT . (2008). The rational exploration of microbial diversity. ISME J 2: 997–1006.
Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM et al. (2009). Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6: 639–U627.
R Development Core Team (2006). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290.
Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S et al. (2010). Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res 38: 5919–5928.
Schutte UME, Abdo Z, Foster J, Ravel J, Bunge J, Solheim B et al. (2010). Bacterial diversity in a glacier foreland of the high Arctic. Mol Ecol 19: 54–66.
Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, Woolf B et al. (2008). Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res 18: 1638–1642.
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Stackebrandt E, Goebel BM . (1994). A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int J Syst Microbiol 44: 846–849.
Suzuki MT, Giovannoni SJ . (1996). Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62: 625–630.
Teixeira L, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J et al. (2010). Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4: 989–1001.
Torsvik V, Daae FL, Sandaa RA, Ovreas L . (1998). Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol 64: 53–62.
Torsvik V, Ovreas L . (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5: 240–245.
Torsvik V, Sorheim R, Goksoyr J . (1996). Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol 17: 170–178.
Uroz S, Buee M, Murat C, Frey-Klett P, Martin F . (2010). Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Reports 2: 281–288.
Vaishampayan P, Osman S, Andersen G, Venkateswaran K . (2010). High-density 16S microarray and clone library-based microbial community composition of the phoenix spacecraft assembly clean room. Astrobiology 10: 499–508.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS . (2009). Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75: 5227–5236.
Zhou JZ . (2009). Predictive microbial ecology. Microb Biotechnol 2: 154–156.
Zhou JZ, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322.
Zhou JZ, Kang S, Schadt CW, Garten CT . (2008). Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA 105: 7768–7773.
Zhou JZ, Thompson DK, Xu Y, Tiedje JM . (2004). Microbial Functional Geomics. John Wiley & Sons: Hoboken, NJ.
Acknowledgements
We thank Dr Fares Najar and Dr Bruce Roe at the University of Oklahoma for providing sequencing services, and Qiong Wang and James Cole at the Michigan State University for helping process pyrosequencing data. This work has been supported, through contract DE-AC02-05CH11231 (as part of ENIGMA, a Scientific Focus Area) and contract DE-SC0004601, by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics: GTL Foundational Science, the United States Department of Agriculture (Project 2007-35319-18305) through NSF-USDA Microbial Observatories Program, Oklahoma Bioenergy Center (OBC) and The Fundamental Research Foundation of Tsinghua University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhou, J., Wu, L., Deng, Y. et al. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5, 1303–1313 (2011). https://doi.org/10.1038/ismej.2011.11
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.11
Keywords
This article is cited by
-
High genome heterozygosity revealed vegetative propagation over the sea in Moso bamboo
BMC Genomics (2023)
-
Natural selenium stress influences the changes of antibiotic resistome in seleniferous forest soils
Environmental Microbiome (2022)
-
Application of Pig Manure Compost with Different Biochar Modifies the Antibiotic Resistome and Bacterial Community in Agriculture Soil
Water, Air, & Soil Pollution (2022)
-
Assessment of microbial α-diversity in one meter squared topsoil
Soil Ecology Letters (2022)
-
Abundance, Diversity and Functional Potentials of Planktonic Bacteria and Microeukaryotes in the Coral-Reef System of Xisha Islands, China
Journal of Ocean University of China (2022)