Abstract
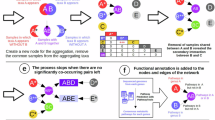
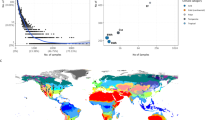
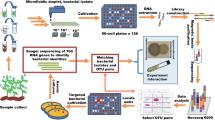
Exploring large environmental datasets generated by high-throughput DNA sequencing technologies requires new analytical approaches to move beyond the basic inventory descriptions of the composition and diversity of natural microbial communities. In order to investigate potential interactions between microbial taxa, network analysis of significant taxon co-occurrence patterns may help to decipher the structure of complex microbial communities across spatial or temporal gradients. Here, we calculated associations between microbial taxa and applied network analysis approaches to a 16S rRNA gene barcoded pyrosequencing dataset containing >160 000 bacterial and archaeal sequences from 151 soil samples from a broad range of ecosystem types. We described the topology of the resulting network and defined operational taxonomic unit categories based on abundance and occupancy (that is, habitat generalists and habitat specialists). Co-occurrence patterns were readily revealed, including general non-random association, common life history strategies at broad taxonomic levels and unexpected relationships between community members. Overall, we demonstrated the potential of exploring inter-taxa correlations to gain a more integrated understanding of microbial community structure and the ecological rules guiding community assembly.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Auguet JC, Barberán A, Casamayor EO . (2010). Global ecological patterns in uncultured Archaea. ISME J 4: 182–190.
Barberán A, Casamayor EO . (2010). Global phylogenetic community structure and beta-diversity patterns of surface bacterioplankton metacommunities. Aquat Microb Ecol 59: 1–10.
Barberán A, Casamayor EO . (2011). Euxinic freshwater hypolimnia promote bacterial endemicity in continental areas. Microb Ecol 61: 465–472.
Bastian M, Heymann S, Jacomy M . (2009). Gephi: An Open Source Software For Exploring and Manipulating Networks. In International AAAI conference on weblogs and social media: San Jose, California.
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N . (2010). Examining the global distribution of dominant archaeal populations in soil. ISME J 5: 908–917.
Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA et al. (2011). The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43: 1450–1455.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Chaffron S, Rehrauer H, Pernthaler J, von Mering C . (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res 20: 947–959.
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R . (2009). Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697.
Csárdi G, Nepusz T . (2006). The igraph software package for complex network research. InterJ Complex Syst 1695: 2006.
Curtis TP, Sloan WT, Scannell JW . (2002). Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA 99: 10494–10499.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626.
Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R et al. (2007). Metagenomic and small-subunit rrna analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol 73: 7059–7066.
Fierer N, Hamady M, Lauber CL, Knight R . (2008). The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA 105: 17994–17999.
Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E . (2010). The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res 38: 3857–3868.
Fuhrman JA, Steele JA . (2008). Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquati Microb Ecol 53: 69–81.
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C . (2009). Unique archaeal assemblages in the arctic ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869.
Guimera R, Amaral LAN . (2005). Functional cartography of complex metabolic networks. Nature 433: 895–900.
Guo Q, Brown JH, Valone TJ . (2000). Abundance and distribution of desert annuals: are spatial and temporal patterns related? J Ecol 88: 551–560.
Holmes AJ, Costello A, Lindstrom ME, Murrell JC . (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbio Lett 132: 203–208.
Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJ, Colwell RK, Fuhrman JA et al. (2007). A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353.
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ . (2001). Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67: 4399–4406.
Janssen PH . (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72: 1719–1728.
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N . (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3: 442–453.
Junker BH, Schreiber F . (2008). Correlation Networks. In: Analysis of Biological Networks. Wiley-Interscience.
Konstantinidis KT, Tiedje JM . (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol 10: 504–509.
Krause AE, Frank KA, Mason DM, Ulanowicz RE, Taylor WW . (2003). Compartments revealed in food-web structure. Nature 426: 282–285.
Lauber CL, Hamady M, Knight R, Fierer N . (2009). Pyrosequencing-based assessment of soil ph as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436.
Magurran AE, Henderson PA . (2003). Explaining the excess of rare species in natural species abundance distributions. Nature 422: 714–716.
McCaig AE, Glover LA, Prosser JI . (2001). Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl Environ Microbiol 67: 4554–4559.
Moody J . (2001). Race, school integration, and friendship segregation in America. Am J Sociol 107: 679–716.
Newman MEJ . (2003). The structure and function of complex networks. SIAM Rev 45: 167–256.
Newman MEJ . (2006). Modularity and community structure in networks. Proc Natl Acad Sci USA 103: 8577.
Oksanen J, Kindt R, Legendre P, O’Hara B, Simpson GL, Solymos P et al. (2007). Vegan: community ecology package. R package version 1: 8.
Pace NR . (1997). A molecular view of microbial diversity and the biosphere. Science 276: 734–740.
Pandit SN, Kolasa J, Cottenie K . (2009). Contrasts between habitat generalists and specialists: an empirical extension to the basic metacommunity framework. Ecology 90: 2253–2262.
Pastor-Satorras R, Vespignani A . (2001). Epidemic spreading in scale-free networks. Phys Rev Lett 86: 3200–3203.
Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, Whitman WB et al. (2010). The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 8: 523–529.
Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP et al. (2007). The role of ecological theory in microbial ecology. Nat Rev Microbiol 5: 384–392.
Proulx SR, Promislow DE, Phillips PC . (2005). Network thinking in ecology and evolution. Trends Ecol Evol 20: 345–353.
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290.
Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F . (2006). Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22: 2532.
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Stone L, Roberts A . (1990). The checkerboard score and species distributions. Oecologia 85: 74–79.
Torsvik V, Goksoyr J, Daae FL . (1990). High diversity in DNA of soil bacteria. Appl Environ Microbiol 56: 782–787.
van der Gast C, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW et al. (2011). Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5: 780–791.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261.
Youssef NH, Elshahed MS . (2009). Diversity rankings among bacterial lineages in soil. ISME J 3: 305–313.
Acknowledgements
We thank members of the Fierer lab, particularly Chris Lauber, Kelly Ramirez and Bob Bowers. We also thank Antoni Fernández-Guerra in the Casamayor lab for QIIME installation and optimization in the CSIC-Blanes server, and members of the Rob Knight lab in UC, particularly Greg Caporaso for computational support and the development of QIIME. AB is supported by the Spanish FPU predoctoral scholarship program and EOC by grants PIRENA CGL2009-13318 and CONSOLIDER-INGENIO 2010 GRACCIE CSD2007-00067 from the Spanish Ministerio de Ciencia e Innovación (MICINN).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Barberán, A., Bates, S., Casamayor, E. et al. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6, 343–351 (2012). https://doi.org/10.1038/ismej.2011.119
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.119
Keywords
This article is cited by
-
Marked variations in diversity and functions of gut microbiota between wild and domestic stag beetle Dorcus Hopei Hopei
BMC Microbiology (2024)
-
Assessing microbiome population dynamics using wild-type isogenic standardized hybrid (WISH)-tags
Nature Microbiology (2024)
-
Network complexity and community composition of key bacterial functional groups promote ecosystem multifunctionality in three temperate steppes of Inner Mongolia
Plant and Soil (2024)
-
Contribution of soil diazotrophs to crop nitrogen utilization in an acidic soil as affected by organic and inorganic amendments
Plant and Soil (2024)
-
Belowground microbiota associated with the progression of Verticillium wilt of smoke trees
Plant and Soil (2024)