Abstract
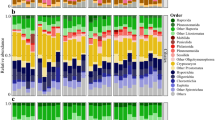
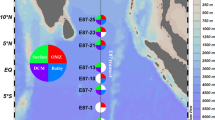
Myxobacteria are common in terrestrial habitats and well known for their formation of fruiting bodies and production of secondary metabolites. We studied a cluster of myxobacteria consisting only of sequences of marine origin (marine myxobacteria cluster, MMC) in sediments of the North Sea. Using a specific PCR, MMC sequences were detected in North Sea sediments down to 2.2 m depth, but not in the limnetic section of the Weser estuary and other freshwater habitats. In the water column, this cluster was only detected on aggregates up to a few meters above the sediment surface, but never in the fraction of free-living bacteria. A quantitative real-time PCR approach revealed that the MMC constituted up to 13% of total bacterial 16S rRNA genes in surface sediments of the North Sea. In a global survey, including sediments from the Mediterranean Sea, the Atlantic, Pacific and Indian Ocean and various climatic regions, the MMC was detected in most samples and to a water depth of 4300 m. Two fosmids of a library from sediment of the southern North Sea containing 16S rRNA genes affiliated with the MMC were sequenced. Both fosmids have a single unlinked 16S rRNA gene and no complete rRNA operon as found in most bacteria. No synteny to other myxobacterial genomes was found. The highest numbers of orthologues for both fosmids were assigned to Sorangium cellulosum and Haliangium ochraceum. Our results show that the MMC is an important and widely distributed but largely unknown component of marine sediment-associated bacterial communities.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. (2000). Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29.
Badger JH, Olsen GJ . (1999). CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol 16: 512–524.
Boyer SL, Flechtner VR, Johansen JR . (2001). Is the 16S–23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol 18: 1057–1069.
Dawid W . (2000). Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev 24: 403–427.
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL . (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27: 4636–4641.
Fieseler L, Quaiser A, Schleper C, Hentschel U . (2006). Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ Microbiol 8: 612–624.
Foudou R, Jojima Y, Yamanaka S . (2002). Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. J Gen Appl Microbiol 48: 109–116.
Garcia R, Gerth K, Stadler M, Dogma IJ, Müller R . (2010). Expanded phylogeny of myxobacteria and evidence for cultivation of the ‘unculturables’. Mol Phylogenet Evol 57: 878–887.
Gerth K, Pradella S, Perlova O, Beyer S, Müller R . (2003). Myxobacteria: proficient producers of novel natural products with various biological activities - past and future biotechnological aspects with the focus on the genus Sorangium. J Biotechnol 106: 233–253.
Giebel HA, Brinkhoff T, Zwisler W, Selje N, Simon M . (2009). Distribution of Roseobacter RCA and SAR11 lineages and distinct bacterial communities from the subtropics to the Southern Ocean. Environ Microbiol 11: 2164–2178.
Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA 100: 8298–8303.
Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA et al. (2006). Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci 103: 15200–15205.
Goodfellow M, Haynes JA . (1984). Actinomycetes in marine sediments. In Ortiz-Ortiz L (ed), Biological, Biochemical and Biomedical Aspects of Actinomycetes. Academic Press: New York, NY, USA, pp 453–472.
Guo FB, Ou HY, Zhang CT . (2003). ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res 31: 1780–1789.
Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H et al. (2004). The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22: 547–553.
Iizuka T, Jojima Y, Fudou R, Hiraishi A, Ahn JW, Yamanaka S . (2003a). Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. J System Evol Microbiol 53: 189–195.
Iizuka T, Jojima Y, Fudou R, Tokura M, Hiraishi A, Yamanaka S . (2003b). Enhygromyxa salina gen. nov., sp. nov., a slightly halophilic Myxobacterium isolated from the coastal areas of Japan. System Appl Microbiol 26: 189–196.
Jago CF, Jones SE, Latter RJ, McCAndliss RR, Hearn MR, Howarth MJ . (2002). Resuspension of benthic fluff by tidal currents in deep stratified waters, northern North Sea. J Sea Res 48: 259–269.
Jiang DM, Kato C, Zhou XW, Wu ZH, Sato T, Li YZ . (2010). Phylogeographic separation of marine and soil myxobacteria at high levels of classification. ISME J 4: 1520–1530.
Köpke B, Wilms R, Engelen B, Cypionka H, Sass H . (2005). Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl Environ Microbiol 71: 7819–7830.
Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA et al. (2009). An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J Biol Chem 284: 9899–9907.
Liesack W, Stackebrandt E . (1989). Evidence for unlinked rrn operons in the planctomycete Pirellula marina. J Bacteriol 171: 5025–5030.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Messing J . (1983). New M13 vectors for cloning. Method Enzymol 101: 20–78.
Mincer TJ, Jensen PR, Kauffman CA, Fenical W . (2002). Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68: 5005–5011.
Mussmann M, Richter M, Lombardot T, Meyerdierks A, Kuever J, Kube M et al. (2005). Clustered genes related to sulfate respiration in uncultured prokaryotes support the theory of their concomitant horizontal transfer. J Bacteriol 187: 7126–7137.
Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C . (1998). Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In Akkermans ADL, van Elsas JD, de Brujin FJ (eds). Molecular Microbial Ecology Manual. 3rd edn Kluwer Academic Publishers: Dordrecht, The Netherlands, pp 1–27, p. 3.4.4.
Muyzer G, Teske A, Wirsen CO, Jannasch HW . (1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis. Arch Microbiol 164: 165–172.
Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M . (2008). GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9: 376.
Overbeek R, Larsen N, Walunas T, D’Souza M, Pusch G, Selkov E et al. (2003). The ERGO genome analysis and discovery system. Nucleic Acids Res 31: 164–171.
Rappé MS, Gordon DA, Vergin KL, Giovannoni SJ . (1999). Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol 22: 106–112.
Reichenbach H . (1999). The ecology of the myxobacteria. Environ Microbiol 1: 15–21.
Reichenbach H . (2005). Bergey's Manual of Systematic Bacteriology 2nd edn, 2: 1059–1144.
Reichenbach H, Dworkin M . (1992). The myxobacteria. In Balows A et al. (eds). The Prokaryotes. 2nd edn, Vol 4. Springer Verlag: New York, pp 3416–3487.
Rink B, Seeberger S, Martens T, Duerselen CD, Simon M, Brinkhoff T . (2007). Effects of a phytoplankton bloom in a coastal ecosystem on the composition of bacterial communities. Aquat Microb Ecol 48: 47–60.
Rückert G . (1984). Untersuchungen zum Vorkommen von Myxobakterien in von Meerwasser beeinflußten Substraten unter besonderer Berücksichtigung der Insel Helgoland. Helgoländer Meeresunters 38: 179–184.
Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes H et al. (2000). The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407: 508–513.
Sanford RA, Cole JR, Tiedje JM . (2002). Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol 68: 893–900.
Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO et al. (2007). Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25: 1281–1289.
Shimkets LJ . (1990). Social and developmental biology of the myxobacteria. Microbiol Rev 54: 473–501.
Shimkets LJ, Dworkin M, Reichenbach H . (2006). The myxobacteria. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds). The Prokaryotes. 3rd edn vol. 7. Springer: Heidelberg, pp 31–115.
Staden R, Beal KF, Bonfield JK . (2000). The Staden package, 1998. Methods Mol Biol 132: 115–130.
Stevens H, Brinkhoff T, Simon M . (2005). Composition and seasonal dynamics of free-living, aggregate- and sediment surface-associated bacterial communities in the German Wadden Sea. Aquat Microb Ecol 38: 15–30.
Süß J, Schubert K, Sass H, Cypionka H, Overmann J, Engelen B . (2006). Widespread distribution and high abundance of Rhizobium radiobacter within Mediterranean subsurface sediments. Environ Microbiol 8: 1753–1763.
Tamas I, Klasson L, Canback B, Naslund AK, Eriksson AS, Wernegreen JJ et al. (2002). 50 million years of genomic stasis in endosymbiotic bacteria. Science 296: 2376–2379.
Tech M, Merkl R . (2003). YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol 3: 441–451.
Wang B, Hu W, Liu H, Zhang CY, Zhao JY, Jiang DM et al. (2007). Adaptation of salt-tolerant Myxococcus strains and their motility systems to the ocean conditions. Microb Ecol 54: 43–51.
Weissman KJ, Müller R . (2010). Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep 27: 1276–1295.
Wenzel SC, Müller R . (2007). Myxobacterial natural product assembly lines: fascinating examples of curious biochemistry. Nat Prod Rep 24: 1211–1224.
Wilms R, Köpke B, Sass H, Chang TS, Cypionka H, Engelen B . (2006). Deep biosphere-related bacteria within the subsurface of tidal flat sediments. Environ Microbiol 8: 709–719.
Wu M, Sun L, Vamathevan J, Riegler M, DeBoy RT, Brownlie J et al. (2003). Direct submission to the NCBI database (www document) http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=383.
Zhang YQ, Li YZ, Wang B, Wu ZH, Zhang CY, Gong X et al. (2005). Characteristics and living patterns of marine myxobacterial isolates. Appl Environ Microbiol 71: 3331–3336.
Zhou J, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322.
Zobell CE, Upham HC . (1944). A list of marine bacteria including descriptions of sixty new species. Bull Scripps Inst Oceanogr Univ Calif 5: 239–292.
Acknowledgements
We thank the crew of the RV Heincke for their valuable support on shipboard, Thomas Badewien and Axel Braun for technical assistance in sampling and Helge Giebel for help with graphical presentations. Furthermore, we thank numerous colleagues and friends for providing samples from various locations. This work was supported by a grant from the Deutsche Forschungsgemeinschaft within the Research Unit ‘BioGeoChemistry of the Wadden Sea’ (FG-432 TP-5 and TP-B).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Brinkhoff, T., Fischer, D., Vollmers, J. et al. Biogeography and phylogenetic diversity of a cluster of exclusively marine myxobacteria. ISME J 6, 1260–1272 (2012). https://doi.org/10.1038/ismej.2011.190
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.190
Keywords
This article is cited by
-
Diversity of Myxobacteria Isolated from Indonesian Mangroves and Their Potential for New Antimicrobial Sources
Current Microbiology (2023)
-
Analysis of the Genome and Metabolome of Marine Myxobacteria Reveals High Potential for Biosynthesis of Novel Specialized Metabolites
Scientific Reports (2018)
-
Current trends in myxobacteria research
Annals of Microbiology (2016)
-
Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone
The ISME Journal (2014)