Abstract
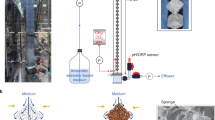
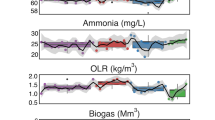
Microbial methanogenesis in subseafloor sediments is a key process in the carbon cycle on the Earth. However, the cultivation-dependent evidences have been poorly demonstrated. Here we report the cultivation of a methanogenic microbial consortium from subseafloor sediments using a continuous-flow-type bioreactor with polyurethane sponges as microbial habitats, called down-flow hanging sponge (DHS) reactor. We anaerobically incubated methane-rich core sediments collected from off Shimokita Peninsula, Japan, for 826 days in the reactor at 10 °C. Synthetic seawater supplemented with glucose, yeast extract, acetate and propionate as potential energy sources was provided into the reactor. After 289 days of operation, microbiological methane production became evident. Fluorescence in situ hybridization analysis revealed the presence of metabolically active microbial cells with various morphologies in the reactor. DNA- and RNA-based phylogenetic analyses targeting 16S rRNA indicated the successful growth of phylogenetically diverse microbial components during cultivation in the reactor. Most of the phylotypes in the reactor, once it made methane, were more closely related to culture sequences than to the subsurface environmental sequence. Potentially methanogenic phylotypes related to the genera Methanobacterium, Methanococcoides and Methanosarcina were predominantly detected concomitantly with methane production, while uncultured archaeal phylotypes were also detected. Using the methanogenic community enrichment as subsequent inocula, traditional batch-type cultivations led to the successful isolation of several anaerobic microbes including those methanogens. Our results substantiate that the DHS bioreactor is a useful system for the enrichment of numerous fastidious microbes from subseafloor sediments and will enable the physiological and ecological characterization of pure cultures of previously uncultivated subseafloor microbial life.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Agrawal L, Ohashi Y, Mochida E, Okui H, Ueki Y, Harada H et al. (1997). Treatment of raw sewage in a temperate climate using a UASB reactor and the hanging sponge cubes process. Wat Sci Technol 36: 433–440.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Aoike K, CK06-06 Scientists. (2007). D/V Chikyu Cruise Offshore Shimokita Laboratory Operation Report. Center for Deep Earth Exploration, JAMSTEC: Yokohama, Japan, http://sio7.jamstec.go.jp/JAMSTEC-exp-report/.
Colwell FS, Boyd S, Delwiche ME, Reed DW, Phelps TJ, Newby DT . (2008). Estimates of biogenic methane production rates in deep marine sediments at Hydrate Ridge, Cascadia Margin. Appl Environ Microbiol 74: 3444–3452.
D'Hondt S, Jørgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA et al. (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306: 2216–2221.
Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G . (2008). Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol 66: 181–196.
Girguis PR, Cozen AE, DeLong EF . (2005). Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl Environ Microbiol 71: 3725–3733.
Girguis PR, Orphan VJ, Hallam SJ, DeLong EF . (2003). Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl Environ Microbiol 69: 5472–5482.
Hattori S . (2008). Syntrophic acetate-oxidizing microbes in methanogenic environments. Microb Environ 23: 118–127.
Imachi H, Sakai S, Hirayama H, Nakagawa S, Nunoura T, Takai K et al. (2008). Exilispira thermophila gen. nov., sp. nov., an anaerobic, thermophilic spirochaete isolated from a deep-sea hydrothermal vent chimney. Int J Syst Evol Microbiol 58: 2258–2265.
Imachi H, Sakai S, Nagai H, Yamaguchi T, Takai K . (2009). Methanofollis ethanolicus sp. nov., an ethanol-utilizing methanogen isolated from a lotus field. Int J Syst Evol Microbiol 59: 800–805.
Imachi H, Sakai S, Ohashi A, Harada H, Hanada S, Kamagata Y et al. (2007). Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int J Syst Evol Microbiol 57: 1487–1492.
Imachi H, Sekiguchi Y, Kamagata Y, Loy A, Qiu YL, Hugenholtz P et al. (2006). Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol 72: 2080–2091.
Inagaki F . (2010). Deep subseafloor microbial community. In: Encyclopedia of Life Sciences. Winley & Sons: Chichester, United Kingdom, doi: 10.1002/9780470015902.a0021894.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA 103: 2815–2820.
Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl Environ Microbiol 69: 7224–7235.
Kendall MM, Boone DR . (2006). Cultivation of methanogens from shallow marine sediments at Hydrate Ridge, Oregon. Archaea 2: 31–38.
Kendall MM, Liu Y, Sieprawska-Lupa M, Stetter KO, Whitman WB, Boone DR . (2006). Methanococcus aeolicus sp. nov., a mesophilic, methanogenic archaeon from shallow and deep marine sediments. Int J Syst Evol Microbiol 56: 1525–1529.
Kobayashi T, Koide O, Mori K, Shimamura S, Matsuura T, Miura T et al. (2008). Phylogenetic and enzymatic diversity of deep subseafloor aerobic microorganisms in organics- and methane-rich sediments off Shimokita Peninsula. Extremophiles 12: 519–527.
Kubota K, Ohashi A, Imachi H, Harada H . (2006). Visualization of mcr mRNA in a methanogen by fluorescence in situ hybridization with an oligonucleotide probe and two-pass tyramide signal amplification (two-pass TSA-FISH). J Microbiol Methods 66: 521–528.
Kvenvolden K . (1995). A review of the geochemistry of methane in natural gas hydrate. Org Geochem 23: 997–1008.
Lipp JS, Morono Y, Inagaki F, Hinrichs K-U . (2008). Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454: 991–994.
Liu Y, Whitman WB . (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea. Ann NY Acad Sci 1125: 171–189.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Marchesi J, Weightman A, Cragg B, Parkes R, Fry J . (2001). Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol Ecol 34: 221–228.
Masui N, Morono Y, Inagaki F . (2008). Microbiological assessment of circulation mud fluids during the first operation of riser drilling by the deep-earth research vessel Chikyu. Geomicrobiol J 25: 274–282.
Meulepas RJW, Jagersma CG, Gieteling J, Buisman CJN, Stams AJM, Lens PNL . (2009). Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol Bioeng 104: 458–470.
Mikucki JA, Liu Y, Delwiche M, Colwell FS, Boone DR . (2003). Isolation of a methanogen from deep marine sediments that contain methane hydrates, and description of Methanoculleus submarinus sp. nov. Appl Environ Microbiol 69: 3311–3316.
Milkov A . (2004). Global estimates of hydrate-bound gas in marine sediments: How much is really out there? Earth Sci Rev 66: 183–197.
Newberry CJ, Webster G, Cragg BA, Parkes RJ, Weightman AJ, Fry JC . (2004). Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ Microbiol 6: 274–287.
Overpeck J, Cole J . (2006). Abrupt change in Earth's climate system. Annu Rev Environ Resour 31: 1–31.
Schink B . (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61: 262–280.
Schoell M . (1988). Multiple origins of methane in the earth. Chem Geol 71: 1–10.
Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA . (2002). Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ Microbiol 4: 713–720.
Stams AJM, Plugge CM . (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7: 568–577.
Tandukar M, Uemura S, Ohashi A, Harada H . (2006). Combining UASB and the ‘fourth generation’ down-flow hanging sponge reactor for municipal wastewater treatment. Wat Sci Technol 53: 209–218.
Toffin L, Webster G, Weightman A, Fry J, Prieur D . (2004). Molecular monitoring of culturable bacteria from deep-sea sediment of the Nankai Trough, Leg 190 Ocean Drilling Program. FEMS Microbiol Ecol 48: 357–367.
Tomaru H, Fehn U, Lu Z, Takeuchi R, Inagaki F, Imachi H et al. (2009). Dating of dissolved iodine in pore waters from the gas hydrate occurrence offshore Shimokita Peninsula, Japan: 129I results from the D/V Chikyu shakedown cruise. Resour Geol 59: 359–373.
Uemura S, Harada H . (2010). Application of UASB technology for sewage treatment with a novel post-treatment process. In: Fang HHP (ed). Environmental Aanaerobic Technology. Imperial College Press: London, United Kingdom, pp 91–112.
von Klein D, Arab H, Völker H, Thomm M . (2002). Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6: 103–110.
von Wintzingerode F, Göbel UB, Stackebrandt E . (1997). Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21: 213–229.
Webster G, Blazejak A, Cragg BA, Schippers A, Sass H, Rinna J et al. (2009). Subsurface microbiology and biogeochemistry of a deep, cold-water carbonate mound from the Porcupine Seabight (IODP Expedition 307). Environ Microbiol 11: 239–257.
Whiticar M . (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161: 291–314.
Zhang Y, Henriet J-P, Bursens J, Boon N . (2010). Stimulation of in vitro anaerobic oxidation of methane rate in a continuous high-pressure bioreactor. Bioresour Technol 101: 3132–3138.
Acknowledgements
We thank Yoshiaki Takahashi (Alt Associates, Ltd, Nagaoka, Japan) for constructing of the DHS reactor; Ai Miyashita, Yuto Yashiro, Masyuki Ehara and Dr Sanae Sakai for assistance with the DHS reactor operation; Takuya Kazawa and Taketoshi Nakamura for help in measuring organic substances; Takeshi Terada for help with Q-PCR; Drs Haruhiko Sumino and Masanobu Takahashi for providing valuable information on the DHS reactor; and Mandy Cumpston, Drs Takuro Nunoura, Satoshi Nakagawa and Hisako Hirayama for helpful discussions and comments. We greatly appreciate Professors Hideki Harada and Akiyoshi Ohashi for their continuous encouragement and support. We also thank the shipboard scientists and crews of the Chikyu Shakedown Expedition CK06-06 for helping us to collect the sediment core sample. This study was partly supported by grants from the Institute of Fermentation, Osaka, Japan, the Japan Society for the Promotion of Science, and the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Imachi, H., Aoi, K., Tasumi, E. et al. Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J 5, 1913–1925 (2011). https://doi.org/10.1038/ismej.2011.64
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.64
Keywords
This article is cited by
-
Cultivation and biogeochemical analyses reveal insights into methanogenesis in deep subseafloor sediment at a biogenic gas hydrate site
The ISME Journal (2022)
-
Isolation of an archaeon at the prokaryote–eukaryote interface
Nature (2020)
-
The effect of carrier addition on Anammox start-up and microbial community: a review
Reviews in Environmental Science and Bio/Technology (2020)
-
Cultivable microbial community in 2-km-deep, 20-million-year-old subseafloor coalbeds through ~1000 days anaerobic bioreactor cultivation
Scientific Reports (2019)
-
Environmental factors shaping the archaeal community structure and ether lipid distribution in a subtropic river and estuary, China
Applied Microbiology and Biotechnology (2018)