Abstract
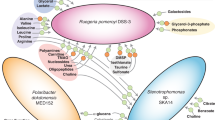
In terms of lifestyle, free-living bacteria are classified as either oligotrophic/specialist or opportunist/generalist. Heterogeneous marine environments such as coastal waters favour the establishment of marine generalist bacteria, which code for a large pool of functions. This is basically foreseen to cope with the heterogeneity of organic matter supplied to these systems. Nevertheless, it is not known what fraction of a generalist proteome is needed for house-keeping functions or what fraction is modified to cope with environmental changes. Here, we used high-throughput proteomics to define the proteome of Ruegeria pomeroyi DSS-3, a model marine generalist bacterium of the Roseobacter clade. We evaluated its genome expression under several natural environmental conditions, revealing the versatility of the bacterium to adapt to anthropogenic influence, poor nutrient concentrations or the presence of the natural microbial community. We also assayed 30 different laboratory incubations to increase proteome coverage and to dig further into the functional genomics of the bacterium. We established its core proteome and the proteome devoted to adaptation to general cellular physiological variations (almost 50%). We suggest that the other half of its theoretical proteome is the opportunist genetic pool devoted exclusively to very specific environmental conditions.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Baudet M, Ortet P, Gaillard JC, Fernandez B, Guerin P, Enjalbal C et al. (2010). Proteomics-based refinement of Deinococcus deserti genome annotation reveals an unwonted use of non-canonical translation initiation codons. Mol Cell Proteomics 9: 415–426.
Biers EJ, Sun S, Howard EC . (2009). Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl Environ Microbiol 75: 2221–2229.
Buchan A, Gonzalez JM, Moran MA . (2005). Overview of the marine roseobacter lineage. Appl Environ Microbiol 71: 5665–5677.
Burgmann H, Howard EC, Ye W, Sun F, Sun S, Napierala S et al. (2007). Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP). Environ Microbiol 9: 2742–2755.
Carvalho PC, Fischer JS, Chen EI, Yates JR, Barbosa VC . (2008). PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics 9: 316.
Christie-Oleza JA, Armengaud J . (2010). In-depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography-tandem mass spectrometry: the Ruegeria pomeroyi DSS-3 case-study. Mar Drugs 8: 2223–2239.
Clair G, Roussi S, Armengaud J, Duport C . (2010). Expanding the known repertoire of virulence factors produced by Bacillus cereus through early secretome profiling in three redox conditions. Mol Cell Proteomics 9: 1486–1498.
de Groot A, Dulermo R, Ortet P, Blanchard L, Guerin P, Fernandez B et al. (2009). Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet 5: e1000434.
DeLong EF . (2009). The microbial ocean from genomes to biomes. Nature 459: 200–206.
Fuhrman JA . (2009). Microbial community structure and its functional implications. Nature 459: 193–199.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M et al. (2006). Essential genes of a minimal bacterium. Proc Natl Acad Sci USA 103: 425–430.
Gupta N, Benhamida J, Bhargava V, Goodman D, Kain E, Kerman I et al. (2008). Comparative proteogenomics: combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res 18: 1133–1142.
Gustavsson N, Diez A, Nystrom T . (2002). The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol Microbiol 43: 107–117.
Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J et al. (2004). The complete genome and proteome of Mycoplasma mobile. Genome Res 14: 1447–1461.
Kato JY, Funa N, Watanabe H, Ohnishi Y, Horinouchi S . (2007). Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc Natl Acad Sci USA 104: 2378–2383.
Keller M, Hettich R . (2009). Environmental proteomics: a paradigm shift in characterizing microbial activities at the molecular level. Microbiol Mol Biol Rev 73: 62–70.
Koonin EV, Wolf YI . (2008). Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36: 6688–6719.
Lanfranconi MP, Bosch R, Nogales B . (2010). Short-term changes in the composition of active marine bacterial assemblages in response to diesel oil pollution. Microbial Biotechnol 3: 607–621.
Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533.
Liu H, Sadygov RG, Yates 3rd JR . (2004). A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201.
Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913.
Moran MA, Belas R, Schell MA, Gonzalez JM, Sun F, Sun S et al. (2007). Ecological genomics of marine Roseobacters. Appl Environ Microbiol 73: 4559–4569.
Mou X, Sun S, Edwards RA, Hodson RE, Moran MA . (2008). Bacterial carbon processing by generalist species in the coastal ocean. Nature 451: 708–711.
Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J 4: 784–798.
Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC et al. (2006). Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci USA 103: 18928–18933.
Polz MF, Hunt DE, Preheim SP, Weinreich DM . (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philosophical Transactions of the Royal Society B-Biological Sciences 361: 2009–2021.
Ramos JL, Gallegos MT, Marques S, Ramos-Gonzalez MI, Espinosa-Urgel M, Segura A . (2001). Responses of Gram-negative bacteria to certain environmental stressors. Curr Opin Microbiol 4: 166–171.
Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299.
Singer E, Webb EA, Nelson WC, Heidelberg JF, Ivanova N, Pati A et al. (2011). The genomic potential of Marinobacter aquaeolei—A biogeochemical opportunitroph. Appl Environ Microbiol 77: 2763–2771.
Sowell SM, Norbeck AD, Lipton MS, Nicora CD, Callister SJ, Smith RD et al. (2008). Proteomic analysis of stationary phase in the marine bacterium ‘Candidatus Pelagibacter ubique’. Appl Environ Microbiol 74: 4091–4100.
Strom SL . (2008). Microbial ecology of ocean biogeochemistry: a community perspective. Science 320: 1043–1045.
Tang K, Huang H, Jiao N, Wu CH . (2010). Phylogenomic analysis of marine Roseobacters. PLoS One 5: e11604.
Ting L, Cowley MJ, Hoon SL, Guilhaus M, Raftery MJ, Cavicchioli R . (2009). Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol Cell Proteomics 8: 2227–2242.
VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF . (2009). Systems biology: Functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol 7: 196–205.
Wagner-Dobler I, Biebl H . (2006). Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60: 255–280.
Wegener KM, Singh AK, Jacobs JM, Elvitigala TR, Welsh EA, Keren N et al. (2010). Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol Cell Proteomics 9: 2678–2689.
Wilmes P, Bond PL . (2006). Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol 14: 92–97.
Wilmes P, Bond PL . (2009). Microbial community proteomics: elucidating the catalysts and metabolic mechanisms that drive the Earth's biogeochemical cycles. Curr Opin Microbiol 12: 310–317.
York GM, Junker BH, Stubbe J, Sinskey AJ . (2001). Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J Bacteriol 183: 4217–4226.
Zech H, Thole S, Schreiber K, Kalhofer D, Voget S, Brinkhoff T et al. (2009). Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 9: 3677–3697.
Acknowledgements
JA Christie-Oleza was supported by a fellowship from the Fundación Ramón Areces. We thank the Commissariat à l’Energie Atomique et aux Energies Alternatives, the Fundación Ramón Areces and the Ministère des Affaires Etrangères et Européennes, République Française (Partenariat Hubert Curien, Picasso Program) for financial support. We also thank C Bruley (CEA-Grenoble) for providing the IRMa parser, Jean-Charles Gaillard and Olivier Pible (both from CEA-Marcoule) for help with data treatment. B Nogales and R Bosch acknowledge the financial support of the Spanish MICINN through project CTM2008-02574/MAR (with FEDER co-funding) and the Acciones Integradas program (FR2009-0106).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Christie-Oleza, J., Fernandez, B., Nogales, B. et al. Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J 6, 124–135 (2012). https://doi.org/10.1038/ismej.2011.86
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.86
Keywords
This article is cited by
-
Microbial oxidation of atmospheric trace gases
Nature Reviews Microbiology (2022)
-
Recombinant myelin oligodendrocyte glycoprotein quality modifies evolution of experimental autoimmune encephalitis in macaques
Laboratory Investigation (2021)
-
Metaproteomics reveal that rapid perturbations in organic matter prioritize functional restructuring over taxonomy in western Arctic Ocean microbiomes
The ISME Journal (2020)
-
Early Colonization of Weathered Polyethylene by Distinct Bacteria in Marine Coastal Seawater
Microbial Ecology (2020)
-
Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide
The ISME Journal (2019)