Abstract
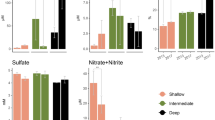
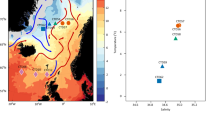
Prokaryote communities were investigated on the seasonally stratified Alaska Beaufort Shelf (ABS). Water and sediment directly underlying water with origin in the Arctic, Pacific or Atlantic oceans were analyzed by pyrosequencing and length heterogeneity-PCR in conjunction with physicochemical and geographic distance data to determine what features structure ABS microbiomes. Distinct bacterial communities were evident in all water masses. Alphaproteobacteria explained similarity in Arctic surface water and Pacific derived water. Deltaproteobacteria were abundant in Atlantic origin water and drove similarity among samples. Most archaeal sequences in water were related to unclassified marine Euryarchaeota. Sediment communities influenced by Pacific and Atlantic water were distinct from each other and pelagic communities. Firmicutes and Chloroflexi were abundant in sediment, although their distribution varied in Atlantic and Pacific influenced sites. Thermoprotei dominated archaea in Pacific influenced sediments and Methanomicrobia dominated in methane-containing Atlantic influenced sediments. Length heterogeneity-PCR data from this study were analyzed with data from methane-containing sediments in other regions. Pacific influenced ABS sediments clustered with Pacific sites from New Zealand and Chilean coastal margins. Atlantic influenced ABS sediments formed another distinct cluster. Density and salinity were significant structuring features on pelagic communities. Porosity co-varied with benthic community structure across sites and methane did not. This study indicates that the origin of water overlying sediments shapes benthic communities locally and globally and that hydrography exerts greater influence on microbial community structure than the availability of methane.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Agogue H, Lamy D, Neal PR, Sogin ML, Herndl GJ . (2011). Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20: 258–274.
Auguet JC, Barberan A, Casamayor EO . (2010). Global ecological patterns in uncultured Archaea. ISME J 4: 182–190.
Baas-Becking LGM . (1934) Geobiologie of inleiding tot de milieukunde (Geobiology as Introduction for Environmental Research) Vol. 18/19. W. P. van Stockum & Zoon: The Hague, Netherlands, p263.
Bano N, Ruffin S, Ransom B, Hollibaugh JT . (2004). Phylogenetic Composition of Arctic Ocean Archaeal Assemblages and Comparison with Antarctic Assemblages. Appl Environ Microbiol 70: 781–789.
Battistuzzi F, Feijao A, Hedges SB . (2004). A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evolutionary Biol 4: 44.
Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA . (2012). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093.
Carmack EC, Macdonald RW . (2002). Oceanography of the Canadian shelf of the Beaufort Sea: a setting for marine life. Arctic 55: 29–45.
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE . (2004). Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70: 1494–1505.
Darby DA, Ortiz J, Polyak L, Lund S, Jakobsson M, Woodgate RA . (2009). The role of currents and sea ice in both slowly deposited central Arctic and rapidly deposited Chukchi-Alaskan margin sediments. Glob Planet Change 68: 56–70.
Dougherty MR . (2012) Compound Specific Carbon Isotope Analysis for Biomarkers Associated with Methanotrophy in the Arctic. Doctoral Dissertation. Department of Chemistry and Biochemistry. University of Maryland: College Park.
Fenchel T . (2003). Biogeography for bacteria. Science 301: 925–926.
Finlay BJ . (2002). Global dispersal of free-living microbial eukaryote species. Science 296: 1061–1063.
Follows MJ, Dutkiewicz S, Grant S, Chisholm SW . (2007). Emergent biogeography of microbial communities in a model ocean. Science 315: 1843–1846.
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C . (2009). Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869.
Galand PE, Lovejoy C, Vincent WF . (2006). Remarkably diverse and contrasting archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat Microb Ecol 44: 115–126.
Galand PE, Potvin M, Casamayor EO, Lovejoy C . (2010). Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J 4: 564–576.
Garneau ME, Vincent WF, Terrado R, Lovejoy C . (2009). Importance of particle-associated bacterial heterotrophy in a coastal Arctic ecosystem. J Marine Syst 75: 185–197.
Grasshoff K, Ehrhardt M, Kremling K, Almgren T . (1983) Methods of Seawater Analysis 2nd rev. and extended edn Verlag Chemie: Weinheim, xxviii, p419.
Griffith DR, Martin WR, Eglinton TI . (2010). The radiocarbon age of organic carbon in marine surface sediments. Geochim Cosmochim Acta 74: 6788–6800.
Hamdan LJ, Gillevet PM, Pohlman JW, Sikaroodi M, Greinert J, Coffin RB . (2011). Diversity and biogeochemical structuring of bacterial communities across the Porangahau ridge accretionary prism, New Zealand. FEMS Microbiol Ecol 77: 518–532.
Hamdan LJ, Jonas RB . (2006). Seasonal and interannual dynamics of free-living bacterioplankton and microbially labile organic carbon along the salinity gradient of the Potomac River. Estuaries Coasts 29: 40–53.
Hamdan LJ, Sikaroodi M, Gillevet PM . (2012). Bacterial community composition and diversity in methane charged sediments revealed by multitag pyrosequencing. Geomicrobiol J 29: 340–351.
Harrison BK, Zhang H, Berelson W, Orphan VJ . (2009). Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California). Appl Environ Microbiol 75: 1487–1499.
Heijs S, Haese R, van der Wielen P, Forney L, van Elsas J . (2007). Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a mediterranean cold seep. Microb Ecol 53: 384–398.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments, on the Pacific Ocean Margin. Proc Natl Acad Sci USA 103: 2815–2820.
Jonas RB . (1997). Bacteria, dissolved organics and oxygen consumption in salinity stratified Chesapeake Bay, an anoxia paradigm. Am Zool 37: 9.
Kirchman DL, Cottrell MT, Lovejoy C . (2010). The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12: 1132–1143.
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG et al (2011). Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8: 761–U107.
Knittel K, Boetius A . (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63: 311–334.
Kvenvolden KA, Lilley MD, Lorenson TD, Barnes PW, Mclaughlin E . (1993). The Beaufort Sea Continental-Shelf as a Seasonal Source of Atmospheric Methane. Geophys Res Lett 20: 2459–2462.
Liao L, Xu X-w, Wang C-s, Zhang D-s, Wu M . (2009). Bacterial and archaeal communities in the surface sediment from the northern slope of the South China Sea. J Zhejiang University Sci B 10: 890–901.
Lozupone C, Hamady M, Knight R . (2006). UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371.
Martin-Cuadrado AB, Rodriguez-Valera F, Moreira D, Alba JC, Ivars-Martinez E, Henn MR et al (2008). Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J 2: 865–886.
Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al (2006). Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
McGuire AD, Anderson LG, Christensen TR, Dallimore S, Guo LD, Hayes DJ et al (2009). Sensitivity of the carbon cycle in the Arctic to climate change. Ecol Monogr 79: 523–555.
McLaughlin FA, Carmack EC, Macdonald RW, Melling H, Swift JH, Wheeler PA et al (2004). The joint roles of Pacific and Atlantic-origin waters in the Canada Basin, 1997-1998. Deep Sea Res Part I 51: 107–128.
Murphy J, Riley JP . (1962). A modified single solution method for determination of phosphate in natural waters. Anal Chim Acta 26: 31.
Orphan VJ, Hinrichs KU, Ussler Iii W, Paull CK, Taylor LT, Sylva SP et al (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol 67: 1922–1934.
Parkes RJ, Cragg BA, Banning N, Brock F, Webster G, Fry JC et al (2007). Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark). Environ Microbiol 9: 1146–1161.
Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ . (2008). Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci 105: 7052–7057.
Pickart RS . (2004). Shelfbreak circulation in the Alaskan Beaufort Sea: Mean structure and variability. J Geophys Res 109: C04024.
Schauer R, Bienhold C, Ramette A, Harder J . (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J 4: 159–170.
Shimada K, Itoh M, Nishino S, McLaughlin F, Carmack E, Proshutinsky A . (2005). Halocline structure in the Canada basin of the arctic ocean. Geophys Res Lett 32: L03605.
Webster G, Parkes RJ, Fry JC, Weightman AJ . (2004). Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol 70: 5708–5713.
Wegener G, Shovitri M, Knittel K, Niemann H, Hovland M, Boetius A . (2008). Biogeochemical processes and microbial diversity of the Gullfaks and Tommeliten methane seeps (Northern North Sea). Biogeosciences 5: 1127–1144.
Wiesenburg DA, Guinasso NL . (1979). Equilibrium solubilities of methane, carbon-monoxide, and hydrogen in water and sea-water. J Chem Eng Data 24: 356–360.
Acknowledgements
LJH was supported by the Naval Research Laboratory (NRL) Chemistry Division Young Investigator Program. Ship time was funded by the NRL platform support program. TT was supported by the Cluster of Excellence ‘The Future Ocean’ funded by the German Research Foundation. We thank the captain and crew of USCGC Polar Sea and C Verlinden, R Downer and L Bryant for field assistance. We thank R Plummer, D Gustafson, the MITAS shipboard scientific party and the nutrient laboratory at Royal NIOZ for laboratory assistance. We thank M Dougherty, K Grabowski, T Lorenson and W Wood for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Hamdan, L., Coffin, R., Sikaroodi, M. et al. Ocean currents shape the microbiome of Arctic marine sediments. ISME J 7, 685–696 (2013). https://doi.org/10.1038/ismej.2012.143
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.143
Keywords
This article is cited by
-
Exploring bacterial diversity in Arctic fjord sediments: a 16S rRNA–based metabarcoding portrait
Brazilian Journal of Microbiology (2024)
-
Bacterial networks in Atlantic salmon with Piscirickettsiosis
Scientific Reports (2023)
-
Geographic Scale Influences the Interactivities Between Determinism and Stochasticity in the Assembly of Sedimentary Microbial Communities on the South China Sea Shelf
Microbial Ecology (2023)
-
Microbial diversity and community structure in deep-sea sediments of South Indian Ocean
Environmental Science and Pollution Research (2022)
-
Major ocean currents may shape the microbiome of the topshell Phorcus sauciatus in the NE Atlantic Ocean
Scientific Reports (2021)