Abstract
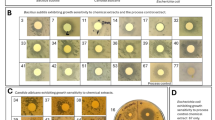
Mono- and multispecies microbial populations alter the chemistry of their surrounding environments during colony development thereby influencing multicellular behavior and interspecies interactions of neighboring microbes. Here we present a methodology that enables the creation of three-dimensional (3D) models of a microbial chemotype that can be correlated to the colony phenotype through multimodal imaging analysis. These models are generated by performing matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) imaging mass spectrometry (IMS) on serial cross-sections of microbial colonies grown on 8 mm deep agar, registering data sets of each serial section in MATLAB to create a model, and then superimposing the model with a photograph of the colonies themselves. As proof-of-principle, 3D models were used to visualize metabolic exchange during microbial interactions between Bacillus subtilis and Streptomyces coelicolor, as well as, Candida albicans and Pseudomonas aeruginosa. The resulting models were able to capture the depth profile of secreted metabolites within the agar medium and revealed properties of certain mass signals that were previously not observable using two-dimensional MALDI-TOF IMS. Most significantly, the 3D models were capable of mapping previously unobserved chemical distributions within the array of sub-surface hyphae of C. albicans and how this chemistry is altered by the presence of P. aeruginosa, an opportunistic pathogen known to alter virulence of C. albicans. It was determined that the presence of C. albicans triggered increased rhamnolipid production by P. aeruginosa, which in turn was capable of inhibiting embedded hyphal growth produced beneath the C. albicans colony at ambient temperature.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Andersson M, Groseclose MR, Deutch AY, Caprioli RM . (2008). Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Meth 5: 101–108.
Bandara HMHN, Yau JYY, Watt RM, Jin LJ, Samaranayake LP . (2010). Pseudomonas aeruginosa inhibits in-vitro Candida biofilm development. BMC Microbiol 10: 125.
Brand A . (2012). Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol 2012: 517529.
Burns JL, Ermerson J, Stapp JR, Yim DL, Krzewinski J, Louden L et al. (1998). Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis 27: 158–163.
Bystrykh LV, Fernandez-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L . (1996). Production of actinorhodin-related ‘blue pigments’ by Streptomyces coelicolor A3(2). J Bacteriol 178: 2238–2244.
Chet I, Mitchell R . (1976). Ecological aspects of microbial chemotactic behavior. Ann Rev Microbiol 30: 221–239.
Chughtai K, Heeren RMA . (2010). Mass spectrometric imaging for biomedical tissue analysis. Chem Rev 110: 3237–3277.
Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B . (2005). Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J Am Soc Mass Spectrom 16: 1093–1099.
Davies J . (2010). How to discover new antibiotics: harvesting the parvome. Curr Opin Chem Biol 15: 5–10.
Davies JC . (2002). Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev 3: 128–134.
Eberlin LS, Ifa DR, Wu C, Cooks RG . (2010). Three-dimensional visualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew Chem 49: 873–876.
Flardh K, Buttner MJ . (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7: 36–49.
Gibson J, Sood A, Hogan DA . (2009). Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. App Environ Microbiol 75: 504–513.
Gonzalez DJ, Haste NM, Hollands A, Fleming TC, Hamby M, Pogliano K et al. (2011). Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 157: 2485–2492.
Hogan DA, Kolter R . (2002). Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296: 2229–2232.
Hogan DA, Vika A, Kolter R . (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54: 1212–1223.
Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Fett F et al. (2002). Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9: 1175–1187.
Kawulka KE, Sprules T, Diaper CM, Whittal RM, McKay RT, Mercier P et al. (2004). Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to alpha-carbon cross-links: formation and reduction of alpha-thio-alpha-amino acid derivatives. Biochemistry 43: 3385–3395.
Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM . (2004). The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene rams in Streptomyces coelicolor. Proc Natl Acad Sci USA 101: 11448–11453.
Liu W-T, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY et al. (2010). Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci USA 107: 16286–16290.
Mcalester G, O’Gara F, Morrissey JP . (2008). Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol 57: 563–569.
McMahon G, Glassner BJ, Lechene CP . (2006). Quantitative imaging of cells with multi-isotope imaging mass spectrometry (MIMS)-Nanoautography with stable isotope tracers. Appl Surf Sci 252: 6895–6906.
Morales DK, Hogan DA . (2010). Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6: e1000886.
Nemes P, Barton AA, Vertes A . (2009). Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal Chem 81: 6668–6675.
Ng W-L, Bassler BL . (2009). Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222.
Phelan VV, Liu W-T, Pogliano K, Dorrestein PC . (2012). Microbial metabolic exchange—the chemotype-phenotype link. Nat Chem Biol 8: 26–35.
Pierce GE . (2005). Pseudomonas aeruginosa, Candida albicans, and device-related nosocomial infections: implications, trends, and potential approaches for control. J Ind Microbiol Biotechnol 32: 309–218.
Romero D, Traxler MF, Lopez D, Kolter R . (2011). Antibiotics as signal molecules. Chem Rev 111: 5492–5505.
Schoenian I, Spiteller M, Ghaste M, Wirth R, Herz H, Spiteller D . (2011). Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci USA 108: 1955–1960.
Schwamborn K, Caprioli RM . (2010). Molecular imaging by mass spectrometry-looking beyond classical histology. Nat Rev Cancer 10: 639–646.
Seeley EH, Caprioli RM . (2012). 3D imaging mass spectrometry: a new frontier. Anal Chem 84: 2105–2110.
Seeley EH, Schwamborn K, Caprioli RM . (2011). Imaging of intact tissue sections: moving beyond the microscope. J Biol Chem 286: 25459–25466.
Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS et al. (2008). Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat Met 5: 57–59.
Soberon-Chavez G, Lepine F, Deziel E . (2005). Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 68: 718–725.
Stanghellini ME, Miller RM . (1997). Biosurfactants: their identity and potential efficacy in the biological control of zoosporic plant pathogens. Plant Dis 81: 4–12.
Straight PD, Willey JM, Kolter R . (2006). Interaction between Streptomyces coelicolor and Bacillus subtilis: role of surfactants in raising aerial structures. J Bacteriol 188: 4918–4925.
Sudbery PE . (2011). Growth of Candida albicans hyphae. Nat Rev Microbiol 9: 737–748.
Truckses DM, Garrenton LS, Thorner J . (2004). Jekyll and Hyde in the microbial world. Science 306: 1509–1511.
Vaidyanathan S, Fletcher JS, Goodacre R, Lockyer NP, Micklefield J, Vickerman JC . (2008). Subsurface biomolecular imaging of Streptomyces coelicolor using secondary ion mass spectrometry. Anal Chem 80: 1942–1951.
Venturi V . (2005). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev 20: 274–291.
Wadhams GH, Armitage JP . (2004). Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cel Biol 5: 1024–1037.
Watrous J, Hendricks N, Meehan M, Dorrestein PC . (2010). Capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Anal Chem 82: 1598–1600.
Watrous JD, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109: 1743–1752.
Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, Fleming TC et al. (2012). A primer to agar-based microbial imaging mass spectrometry. J Bacteriol 194: 6023–6028.
Yang Y-L, Xu Y, Kersten RD, Liu WT, Meehan MJ, Moore BS et al. (2011). Connecting chemotypes and phenotypes of cultured marine microbial assemblages using imaging mass spectrometry. Angew Chem 50: 5839–5842.
Yang Y-L, Xu Y, Straight P, Dorrestein PC . (2009). Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 5: 885–887.
Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T et al. (2008). The origins of 168, W23 and other Bacillus subtilis legacy strains. J Bacteriol 190: 6983–6995.
Zhang S . (2004). Beyond the Petri dish. Nat Biotechnol 22: 151–152.
Acknowledgements
This work was supported by the Keck Foundation and National Institute of General Medical Sciences Grant NIH GM086283, GM094802 (PCD), NIH Molecular Biophysics Training Grant NIH GM08326 (JDW), and European Union 7th Framework Programme grant 305259. We thank Suzanne Noble, University of California San Francisco, for kindly providing us with C. albicans ySN250.
Author contributions
JDW, TA and PCD designed the research plan; JDW, VVP and C-CH performed the experiments; TA performed 3D modeling of the MALDI-TOF IMS data; C-CH, WJM and BMD performed NMR analysis; JDW, VVP, C-CH, WJM and PCD analyzed MS data; JDW, VVP, C-CH, TA and PCD wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Watrous, J., Phelan, V., Hsu, CC. et al. Microbial metabolic exchange in 3D. ISME J 7, 770–780 (2013). https://doi.org/10.1038/ismej.2012.155
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.155
Keywords
This article is cited by
-
Mass spectrometry imaging for biosolids characterization to assess ecological or health risks before reuse
Nature Communications (2023)
-
Three-dimensional (3D) imaging of lipids in skin tissues with infrared matrix-assisted laser desorption electrospray ionization (MALDESI) mass spectrometry
Analytical and Bioanalytical Chemistry (2021)
-
In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids
Nature Microbiology (2019)
-
Observed metabolic asymmetry within soybean root nodules reflects unexpected complexity in rhizobacteria-legume metabolite exchange
The ISME Journal (2018)
-
A Simple Sonication Improves Protein Signal in Matrix-Assisted Laser Desorption Ionization Imaging
Journal of the American Society for Mass Spectrometry (2018)