Abstract
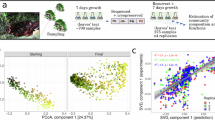
Animals are colonized by complex bacterial communities. The processes controlling community membership and influencing the establishment of the microbial ecosystem during development are poorly understood. Here we aimed to explore the assembly of bacterial communities in Hydra with the broader goal of elucidating the general rules that determine the temporal progression of bacterial colonization of animal epithelia. We profiled the microbial communities in polyps at various time points after hatching in four replicates. The composition and temporal patterns of the bacterial communities were strikingly similar in all replicates. Distinct features included high diversity of community profiles in the first week, a remarkable but transient adult-like profile 2 weeks after hatching, followed by progressive emergence of a stable adult-like pattern characterized by low species diversity and the preponderance of the Betaproteobacterium Curvibacter. Intriguingly, this process displayed important parallels to the assembly of human fecal communities after birth. In addition, a mathematical modeling approach was used to uncover the organizational principles of this colonization process, suggesting that both, local environmental or host-derived factor(s) modulating the colonization rate, as well as frequency-dependent interactions of individual bacterial community members are important aspects in the emergence of a stable bacterial community at the end of development.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ayres JS, Trinidad NJ, Vance RE . (2012). Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med 18: 799–806.
Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H et al. (2009). Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol 33: 559–569.
Bosch TC, McFall-Ngai MJ . (2011). Metaorganisms as the new frontier. Zoology 114: 185–190.
Bosch TC . (2012). Understanding complex host-microbe interactions in Hydra. Gut Microbes 3: 345–351.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T et al. (2010). The dynamic genome of Hydra. Nature 464: 592–596.
Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK . (2010). Host-bacterial symbiosis in health and disease. Adv Immunol 107: 243–274.
Cilieborg MS, Boye M, Sangild PT . (2012). Bacterial colonization and gut development in preterm neonates. Early Hum Dev 88 (Suppl 1): S41–S49.
Drake JA . (1990). The mechanics of community assembly and succession. J Theor Biol 147: 213–233.
Franzenburg S, Fraune S, Künzel S, Baines JF, Domazet-Lôso T, Bosch TC . (2012). MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA 109: 19374–19379.
Fraune S, Abe Y, Bosch TC . (2009). Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ Microbiol 11: 2361–2369.
Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB et al. (2010). In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA 107: 18067–18072.
Fraune S, Bosch TC . (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA 104: 13146–13151.
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504.
Hemmrich G, Anokhin B, Zacharias H, Bosch TC . (2007). Molecular phylogenetics in Hydra, a classical model in evolutionary developmental biology. Mol Phylogenet Evol 44: 281–290.
Hemmrich G, Khalturin K, Boehm AM, Puchert M, Anton-Erxleben F, Wittlieb J et al. (2012). Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol 29: 3267–3280.
Hofbauer J, Sigmund K . (1998) Evolutionary Games and Population Dynamics. Cambridge University Press: Cambridge.
Hooper LV, Gordon JI . (2001). Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11: 1R–10R.
Kelly D, King T, Aminov R . (2007). Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res 622: 58–69.
Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108 (Suppl 1): 4578–4585.
Lange C, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Miller DJ, Rahn T et al. (2011). Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol 28: 1687–1702.
Lenhoff HM, Brown RD . (1970). Mass culture of hydra: an improved method and its application to other aquatic invertebrates. Lab Anim 4: 139–154.
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI . (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788.
Ley RE, Peterson DA, Gordon JI . (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124: 837–848.
Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M et al. (2011). Antimicrobial peptides keep insect endosymbionts under control. Science 334: 362–365.
Mao-Jones J, Ritchie KB, Jones LE, Ellner SP . (2010). How microbial community composition regulates coral disease development. PLoS Biol 8: e1000345.
Martin VJ, Littlefield CL, Archer WE, Bode HR . (1997). Embryogenesis in Hydra. Biol Bull 192: 345–363.
Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL . (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118.
Moran AP, Gupta A, Joshi L . (2011). Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60: 1412–1425.
Murray JD . (2002) Mathematical Biology: I. An Introduction. Springer: Berlin.
Muyzer G, de Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700.
Nyholm SV, McFall-Ngai MJ . (2004). The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2: 632–642.
Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH et al. (2010). Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546.
Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336: 489–493.
Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO . (2007). Development of the human infant intestinal microbiota. PLoS Biol 5: e177.
Rawls JF, Samuel BS, Gordon JI . (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA 101: 4596–4601.
Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11: 76–83.
Sandstrom J, Telang A, Moran NA . (2000). Nutritional enhancement of host plants by aphids – a comparison of three aphid species on grasses. J Insect Physiol 46: 33–40.
Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334: 670–674.
Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV . (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA 105: 20858–20863.
Walter J, Ley R . (2011). The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol 65: 411–429.
Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC . (2006). Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Nat Acad Sci USA 103: 6208–6211.
Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299: 2074–2076.
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) through grant Bo 848/17-1, and grants from the DFG Cluster of Excellence programs ‘The Future Ocean’ and ‘Inflammation at Interfaces’ (to TCGB). PMA and AT acknowledge financial support from the DFG through an Emmy-Noether grant and from the Max Planck Society. We thank Heinke Buhtz for her technical support in the 454 sequencing process.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Franzenburg, S., Fraune, S., Altrock, P. et al. Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J 7, 781–790 (2013). https://doi.org/10.1038/ismej.2012.156
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.156
Keywords
This article is cited by
-
On being a Hydra with, and without, a nervous system: what do neurons add?
Animal Cognition (2023)
-
A combination of host ecology and habitat but not evolutionary history explains differences in the microbiomes associated with rotifers
Hydrobiologia (2023)
-
Beyond Lynn Margulis’ green hydra
Symbiosis (2022)
-
Bdellovibrio and Like Organisms Are Predictors of Microbiome Diversity in Distinct Host Groups
Microbial Ecology (2020)
-
Habitat filters mediate successional trajectories in bacterial communities associated with the striped shore crab
Oecologia (2019)