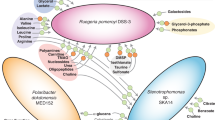
Abstract
Bacteroidetes are commonly assumed to be specialized in degrading high molecular weight (HMW) compounds and to have a preference for growth attached to particles, surfaces or algal cells. The first sequenced genomes of marine Bacteroidetes seemed to confirm this assumption. Many more genomes have been sequenced recently. Here, a comparative analysis of marine Bacteroidetes genomes revealed a life strategy different from those of other important phyla of marine bacterioplankton such as Cyanobacteria and Proteobacteria. Bacteroidetes have many adaptations to grow attached to particles, have the capacity to degrade polymers, including a large number of peptidases, glycoside hydrolases (GHs), glycosyl transferases, adhesion proteins, as well as the genes for gliding motility. Several of the polymer degradation genes are located in close association with genes for TonB-dependent receptors and transducers, suggesting an integrated regulation of adhesion and degradation of polymers. This confirmed the role of this abundant group of marine bacteria as degraders of particulate matter. Marine Bacteroidetes had a significantly larger number of proteases than GHs, while non-marine Bacteroidetes had equal numbers of both. Proteorhodopsin containing Bacteroidetes shared two characteristics: small genome size and a higher number of genes involved in CO2 fixation per Mb. The latter may be important in order to survive when floating freely in the illuminated, but nutrient-poor, ocean surface.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Alonso C, Warnecke F, Amann R, Pernthaler J . (2007). High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol 9: 1253–1266.
Alonso-Sáez L, Gasol J . (2007). Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in Northwestern Mediterranean Coastal waters. Appl Environ Microbiol 73: 3528–3535.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Amaral-Zettler L, Artigas LF, Baross J, Loka Bharathi PA, Boetius A, Chandramohan D et al. (2010). A global census of marine life. In: McIntyre AD, (eds) Life in the World’s Oceans. Blackwell Publishing Ltd.: Chichester, West Sussex, UK, pp. 223–245.
Andersson DI, Hughes D . (2009). Gene amplification and adaptive evolution in bacteria. Annu Rev Genet 43: 167–195.
Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E et al. (2006). Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ Microbiol 8: 2201–2213.
Bendtsen JD, Nielsen H, von Heijne G, Brunak S . (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795.
Castresana J . (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552.
Cottrell MT, Kirchman DL . (2000). Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacteria cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66: 1692–1697.
DeLong EF, Béjà O . (2010). The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol 8: e1000359.
DeLong EF, Franks DG, Alldredge AL . (1993). Phylogenetic diversity of aggregate-attached vs. free-fiving marine macterial assemblages. Limnol Oceanogr 38: 924–934.
Fernández-Gómez B, Fernàndez-Guerra A, Casamayor EO, González JM, Pedrós-Alió C, Acinas SG . (2012). Patterns and architecture of genomic islands in marine bacteria. BMC Genomics 13: 347–366.
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R et al. (2008). The Pfam protein families database. Nucleic Acids Res 36: D281–D288.
Fuhrman JA, Schwalbach MS, Stingl U . (2008). Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol 6: 488–494.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Glöckner FO, Fuchs BM, Amann R . (1999). Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol 65: 3721–3726.
González JM, Pinhassi J, Fernández-Gómez B, Coll-Llado M, González-Velázquez M, Puigbó P et al. (2011). Genomics of the proteorhodopsin-containing marine Flavobacterium Dokdonia sp. strain MED134. Appl Environ Microbiol 77: 8676–8686.
Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL et al. (2010). Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol 8: e1000358.
Gómez-Consarnau L, González JM, Coll-Lladó M, Gourdon P, Pascher T, Neutze R et al. (2007). Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445: 210–213.
Gómez-Pereira PR, Fuchs BM, Alonso C, Oliver M, van Beusekom J, Amann R . (2010). Distribution patterns and diversity of planktonic Flavobacterial clades in contrasting water masses of the North Atlantic Ocean. ISME J 4: 472–487.
Gómez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J et al. (2012). Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ Microbiol 14: 52–66.
Gónzalez JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M et al. (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc Natl Acad Sci USA 105: 8724–8729.
Haft DH, Selengut J, Mongodin EF, Nelson KE . (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60.
Hou S, Saw JH, Lee KS, Freitas TA, Belisle C, Kawarabayasi Y et al. (2004). Genome sequence of the deep-sea gamma-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc Natl Acad Sci USA 101: 18036–18041.
Hsiao WWL, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FSL . (2005). Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genetics 1: 540–550.
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215.
Jarrell KF, McBride MJ . (2008). The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6: 466–476.
Kimura H, Young CR, Martínez A, DeLong EF . (2011). Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J 5: 1641–1651.
Kirchman D . (2002). The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39: 91–100.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL . (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305: 567–580.
Langille MGI, Brinkman FSL . (2009). IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25: 664–665.
Langille MGI, Hsiao WWL, Brinkman FSL . (2010). Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol 8: 373–382.
Letunic I, Bork P . (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128.
Letunic I, Bork P . (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478.
Martens EC, Koropatkin NM, Smith TJ, Gordon JI . (2009). Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284: 24673–24677.
McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG et al. (2009). Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75: 6864–6875.
Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J et al. (2003). GenDB--an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31: 2187–2195.
Pedrotti ML, Beauvais S, Kerros ME, Iversen K, Peters F . (2009). Bacterial colonization of transparent exopolymeric particles in mesocosms under different turbulence intensities and nutrient conditions. Aquat Microb Ecol 55: 301–312.
Pinhassi J, Bowman JP, Nedashkovskaya OI, Lekunberri I, Gómez-Consarnau L, Pedrós-Alió C . (2006). Leeuwenhoekiella blandensis sp. nov., a genome-sequenced marine member of the family Flavobacteriaceae. Int J Sys Evol Microbiol 56: 1489–1493.
Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol 70: 6753–6766.
Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Pushker R, Mira A, Rodríguez-Valera F . (2004). Comparative genomics of gene-family size in closely related bacteria. Genome Biol 5: R27.
Richter M, Lombardot T, Kostadinov I, Kottmann R, Duhaime M, Peplies J et al. (2008). JCoast—A biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes. BMC Bioinformatics 9: 177.
Schut F, de Vries EJ, Gottschal JC, Robertson BR, Harder W, Prins RA et al. (1993). Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol 59: 2150–2160.
Stamatakis A, Hoover P, Rougemont J . (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771.
Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ . (2011). Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS ONE 6: e19725.
Stingl U, Desiderio RA, Cho J-C, Vergin KL, Giovannoni SJ . (2007). The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol 73: 2290–2296.
Tatusov RL, Koonin EV, Lipman DJ . (1997). A genomic perspective on protein families. Science 278: 631–637.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611.
Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H et al. (2009). Assembling the marine metagenome, one cell at a time. PLoS ONE 4: e5299.
Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M et al. (2010). Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468: 60–66.
Acknowledgements
BF-G was a recipient of an I3P grant from CSIC. The genomes of the three MED strains were sequenced by the JCVI through the Marine Microbial Initiative of the Gordon and Betty Moore Foundation. We thank Thierry Lombardot for bioinformatic analysis. This work was supported by grants ‘GEMMA’ CTM2007-63753-C02-01/MAR and ‘BBMOGenome’ CTM2010-11060-E from the Spanish Ministerio de Ciencia e Innovación.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Fernández-Gómez, B., Richter, M., Schüler, M. et al. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J 7, 1026–1037 (2013). https://doi.org/10.1038/ismej.2012.169
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.169
Keywords
This article is cited by
-
Autochthonous psychrophilic hydrocarbonoclastic bacteria and its ecological function in contaminated cold environments
Biodegradation (2024)
-
Deep-sea Bacteroidetes from the Mariana Trench specialize in hemicellulose and pectin degradation typically associated with terrestrial systems
Microbiome (2023)
-
Bacterial response to glucose addition: growth and community structure in seawater microcosms from North Pacific Ocean
Scientific Reports (2023)
-
Metabolic handoffs between multiple symbionts may benefit the deep-sea bathymodioline mussels
ISME Communications (2023)
-
Chemical Links Between Redox Conditions and Estimated Community Proteomes from 16S rRNA and Reference Protein Sequences
Microbial Ecology (2023)