Abstract
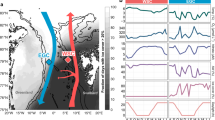
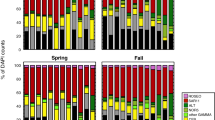
A metaproteomic survey of surface coastal waters near Palmer Station on the Antarctic Peninsula, West Antarctica, was performed, revealing marked differences in the functional capacity of summer and winter communities of bacterioplankton. Proteins from Flavobacteria were more abundant in the summer metaproteome, whereas winter was characterized by proteins from ammonia-oxidizing Marine Group I Crenarchaeota. Proteins prevalent in both seasons were from SAR11 and Rhodobacterales clades of Alphaproteobacteria, as well as many lineages of Gammaproteobacteria. The metaproteome data were used to elucidate the main metabolic and energy generation pathways and transport processes occurring at the microbial level in each season. In summer, autotrophic carbon assimilation appears to be driven by oxygenic photoautotrophy, consistent with high light availability and intensity. In contrast, during the dark polar winter, the metaproteome supported the occurrence of chemolithoautotrophy via the 3-hydroxypropionate/4-hydroxybutyrate cycle and the reverse tricarboxylic acid cycle of ammonia-oxidizing archaea and nitrite-oxidizing bacteria, respectively. Proteins involved in nitrification were also detected in the metaproteome. Taurine appears to be an important source of carbon and nitrogen for heterotrophs (especially SAR11), with transporters and enzymes for taurine uptake and degradation abundant in the metaproteome. Divergent heterotrophic strategies for Alphaproteobacteria and Flavobacteria were indicated by the metaproteome data, with Alphaproteobacteria capturing (by high-affinity transport) and processing labile solutes, and Flavobacteria expressing outer membrane receptors for particle adhesion to facilitate the exploitation of non-labile substrates. TonB-dependent receptors from Gammaproteobacteria and Flavobacteria (particularly in summer) were abundant, indicating that scavenging of substrates was likely an important strategy for these clades of Southern Ocean bacteria. This study provides the first insight into differences in functional processes occurring between summer and winter microbial communities in coastal Antarctic waters, and particularly highlights the important role that ‘dark’ carbon fixation has in winter.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Abell GCJ, Bowman JP . (2005). Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol Ecol 51: 265–277.
Allen M, Lauro FM, Williams TJ, Burg D, Siddiqui KS, De Francisci D et al. (2009). The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold-adaptation. ISME J 3: 1012–1035.
Australian Commonwealth Government Australian Antarctic Science Strategic Plan 2011–12 to 2020–21 (2011). Australian Commonwealth Government: ISBN 978 1 876934 16 6
Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP et al. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 15: 1902–1906.
Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF . (2001). Proteorhodopsin phototrophy in the ocean. Nature 411: 786–789.
Berg IA, Kockelkorn D, Buckel W, Fuchs G . (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318: 1782–1786.
Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N et al. (2007). Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2: 224.
Buchan A, González JM, Moran MA . (2005). Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71: 5665–5677.
Burg DW, Lauro FM, Williams TJ, Raftery MJ, Guilhaus M, Cavicchioli R . (2010). Analyzing the hydrophobic proteome of the antarctic archaeon Methanococcoides burtonii using differential solubility fractionation. J Proteome Res 9: 664–676.
Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF et al. (2010). A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330: 1375–1378.
Cho JC, Giovannoni SJ . (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol 70: 432–440.
Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM . (2003). Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr 48: 1893–1902.
Cottrell MT, Kirchman DL . (2000). Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular weight dissolved organic matter. Appl Environ Microbiol 66: 1692–1697.
Danson MJ, Lamble HJ, Hough DW . (2007). Central metabolism. In: Caviccholi R (ed): Archaea: Molecular and Cellular Biology. ASM Press: Washington DC, pp 260–287.
DeLong EF, Béjà O . (2010). The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol 8: e1000359.
DeLong EF, Wu KY, Prezelin BB, Jovine RVM . (1994). High abundance of Archaea in Antarctic marine picoplankton. Nature 371: 695–697.
DeMaere MZ, Lauro FM, Thomas T, Yau S, Cavicchioli R . (2011). Simple high-throughput annotation pipeline (SHAP). Bioinformatics 27: 2431–2432.
Dixon JL, Beale R, Nightingale PD . (2011). Rapid biological oxidation of methanol in the tropical Atlantic: significance as a microbial carbon source. Biogeosci Discuss 8: 3899–3921.
Farmer CT, Hansell DH . (2007). Determination of dissolved organic carbon and total dissolved nitrogen in sea water. In: Dickson AG, Sabina CL, Christian JR (eds): Guide to Best Practices for Ocean CO2 Measurements PICES Special Publication: Sydney, British Columbia, Canada, p 3191.
Forward JA, Behrendt MC, Wyborn NR, Cross R, Kelly DJ . (1997). TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J Bacteriol 179: 5482–5493.
Fuhrman JA, Schwalbach MS, Stingl U . (2008). Proteorhodopsins: an array of physiological roles?. Nat Rev Microbiol 6: 488–494.
Ghiglione JF, Murray AE . (2012). Pronounced summer to winter differences and higher wintertime richness in coastal sub-Antarctic and Antarctic marine bacterioplankton. Environ Microbiol 14: 617–629.
Gilbert HJ . (2008). Sus out sugars in. Structure 16: 987–989.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL et al. (2005). Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85.
Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782.
Goltsman DS, Denef VJ, Singer SW, VerBerkmoes NC, Lefsrud M et al. (2009). Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (Group II) and “Leptospirillum ferrodiazotrophum” (Group III) bacteria in acid mine drainage biofilms. Appl Environ Microbiol 75: 4599–4615.
Gómez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R et al. (2007). Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445: 210–213.
González JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M et al. (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc Natl Acad Sci USA 105: 8724–8729.
Grossart H-P, Levold F, Allgaier M, Simon M, Brinkhoff T . (2005). Marine diatom species harbour distinct bacterial communities. Environ Microbiol 7: 860–873.
Grzymski JJ, Riesenfeld CS, Williams TJ, Dussaq AM, Ducklow H, Erickson M et al. (2012). A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J; e-pub ahead of print; doi:10.1038/ismej.2012.31.
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J et al. (2006a). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA 103: 18296–18301.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006b). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95.
Hayashi NR, Arai H, Kodama T, Igarashi Y . (1997). The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem Biophys Res Commun 241: 565–569.
Heikes BG, Chang W, Pilson MEQ, Swift E, Singh HB, Guenther A et al. (2002). Atmospheric methanol budget and ocean implication. Global Biogeochem Cycles 16: 1133.
Holtmann G, Bremer E . (2004). Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J Bacteriol 186: 1683–1693.
Kappler U . (2011). Bacterial sulfite-oxidizing enzymes. Biochim Biophys Acta 1807: 1–10.
Karner M, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–509.
Kirchman D, K'Nees E, Hodson R . (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol 49: 599–607.
Kockelkorn D, Fuchs G . (2009). Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J Bacteriol 191: 6352–6362.
Koebnik R . (2005). TonB-dependent trans-envelope signalling: the exception or the rule?. Trends Microbiol 13: 343–347.
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Lauro FM, DeMaere MZ, Yau S, Brown M, Ng C, Wilkins D et al. (2011). An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5: 879–895.
Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533.
Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107: 13479–13484.
Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R et al. (2003). Diversification and spectral tuning in marine proteorhodopsins. EMBO J 22: 1725–1731.
Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Grechkin Y et al. (2012). IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res 40: (Database issue) D123–D129.
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA . (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979.
Massana R, Pedros-Alio C, Casamayor EO, Gasol JM . (2001). Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol Oceanogr 46: 1181–1188.
Meyer B, Kuever J . (2007). Molecular analysis of the diversity of sulfate-reducing and sulfur-oxidizing prokaryotes in the environment, using aprA as functional marker gene. Appl Environ Microbiol 73: 7664–7679.
Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S et al. (2007). Ecological genomics of marine Roseobacters. Appl Environ Microbiol 73: 4559–4569.
Moran MA, Zepp RG . (1997). Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limnol Oceanogr 42: 1307–1316.
Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G . (2010). Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J 4: 673–685.
Murray AE, Grzymski JJ . (2007). Diversity and genomics of Antarctic marine micro-organisms. Philos Trans R Soc Lond B Biol Sci 362: 2259–2271.
Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K et al. (1998). Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters off Anvers Island, Antarctica. Appl Environ Microbiol 64: 2585–2595.
Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F . (2007). Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene deletion mutants. J Med Microbiol 56: 1536–1548.
Newton IL, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC et al. (2007). The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315: 998–1000.
Ng C, DeMaere MZ, Williams TJ, Lauro FM, Raftery M, Gibson JA et al. (2010). Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J 4: 1002–1019.
Piquet AM, Bolhuis H, Meredith MP, Buma AG . (2011). Shifts in coastal Antarctic marine microbial communities during and after melt water-related surface stratification. FEMS Microbiol Ecol 76: 413–427.
Ramos-Vera WH, Weiss M, Strittmatter E, Kockelkorn D, Fuchs G . (2011). Identification of missing genes and enzymes for autotrophic carbon fixation in crenarchaeota. J Bacteriol 193: 1201–1211.
Reeves AR, Wang G-R, Salyers AA . (1997). Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol 179: 643–649.
Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB . (2011). Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473: 208–211.
Rodriguez-Brito B, Rohwer F, Edwards RA . (2006). An application of statistics to comparative metagenomics. BMC Bioinformatics 7: 162.
Sharp JH, Benner R, Carlson CA, Dow R, Fitzwater SE . (1993). Re-evaluation of high temperature combustion and chemical oxidation measurements of dissolved organic carbon in seawater. Limnol Oceanogr 38: 1774–1782.
Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J . (2009). Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol 11: 2078–2093.
Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ . (2010). The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol 12: 490–500.
Smith DP, Kitner JB, Norbeck AD, Clauss TR, Lipton MS, Schwalbach MS et al. (2010). Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus Pelagibacter ubique. PLoS One 5: e10487.
Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF et al. (2011). Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5: 856–865.
Sowell SM, Norbeck AD, Lipton MS, Nicora CD, Callister SJ, Smith RD et al. (2008). Proteomic analysis of stationary phase in the marine bacterium “Candidatus Pelagibacter ubique”. Appl Environ Microbiol 74: 4091–4100.
Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF et al. (2009). Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3: 93–105.
Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ . (2011). Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One 6: e19725.
Stewart FJ, Ulloa O, Delong EF . (2012). Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23–40.
Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF . (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA 105: 4209–4214.
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440: 790–794.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW . (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144.
Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE et al. (2011). One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6: e23973.
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300.
Thomas T, Egan S, Burg D, Ng C, Ting L, Cavicchioli R . (2007). The integration of genomics and proteomics into marine, microbial ecology. Theme series on “Genomics, proteomics and metabolomics in marine ecology”. Mar Ecol Prog Ser 332: 291–299.
Ting L, Williams TJ, Cowley MJ, Lauro FM, Guilhaus M, Raftery MJ et al. (2010). Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ Microbiol 12: 2658–2676.
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425.
Tripp HJ, Kitner JB, Schwalbach MS, Dacey JW, Wilhelm LJ, Giovannoni SJ . (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452: 741–744.
Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ . (2009). Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol 11: 230–238.
Turner J, Colwell SR, Marshall GJ, Lachlan-Cope TA, Carleton AM, Jones PD et al. (2005). Antarctic climate change during the last 50 years. Int J Climatol 25: 279–294.
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823.
Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582.
Williams TJ, Burg DW, Raftery MJ, Poljak A, Guilhaus M, Pilak O et al. (2010). Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii Part I: the effect of growth temperature. J Proteome Res 9: 640–652.
Williams TJ, Ertan H, Ting L, Cavicchioli R . (2009). Carbon and nitrogen substrate utilization in the marine bacterium Sphingopyxis alaskensis strain RB22. ISME J 3: 1036–1052.
Winnen B, Hvorup RN, Saier MH . (2003). The tripartite tricarboxylate transporter (TTT) family. Res Microbiol 154: 457–465.
Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P et al. (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103: 12317–12322.
Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ et al. (2011). Virophage control of Antarctic algal host–virus dynamics. Proc Natl Acad Sci USA 108: 6163–6168.
Acknowledgements
We extend our thanks to our field team M Erickson, K Myers, V Peng and J-F Ghiglione for their expert assistance sampling in Antarctica and to the Palmer Station personnel in the US Antarctic Program for support, especially during the winter field season. The work was supported by the NSF grants, ANT 0632389 to AEM and JJG, and ANT 0632278 and ANT 0217282 to HD. The work of the Australian contingent was supported by the Australian Research Council. Mass spectrometric results were obtained at the Bioanalytical Mass Spectrometry Facility within the Analytical Centre of the University of New South Wales. This work was undertaken using infrastructure provided by NSW Government co-investment in the National Collaborative Research Infrastructure Scheme. Subsidized access to this facility is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Williams, T., Long, E., Evans, F. et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J 6, 1883–1900 (2012). https://doi.org/10.1038/ismej.2012.28
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.28
Keywords
This article is cited by
-
Jellyfish detritus supports niche partitioning and metabolic interactions among pelagic marine bacteria
Microbiome (2023)
-
Seasonal patterns in microbial carbon and iron transporter expression in the Southern Ocean
Microbiome (2023)
-
Vertical microbial profiling of water column reveals prokaryotic communities and distribution features of Antarctic Peninsula
Acta Oceanologica Sinica (2023)
-
The contribution of nirK gene-containing thaumarchaea to denitrification and N2O production across coastal sediment and terrestrial ecosystems
Journal of Soils and Sediments (2022)
-
North Sea spring bloom-associated Gammaproteobacteria fill diverse heterotrophic niches
Environmental Microbiome (2021)