Abstract
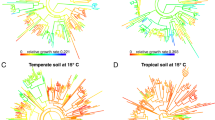
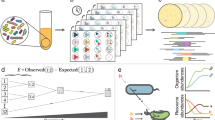
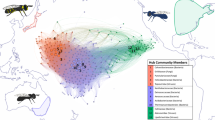
High-throughput sequencing techniques have made large-scale spatial and temporal surveys of microbial communities routine. Gaining insight into microbial diversity requires methods for effectively analyzing and visualizing these extensive data sets. Phylogenetic β-diversity measures address this challenge by allowing the relationship between large numbers of environmental samples to be explored using standard multivariate analysis techniques. Despite the success and widespread use of phylogenetic β-diversity measures, an extensive comparative analysis of these measures has not been performed. Here, we compare 39 measures of phylogenetic β diversity in order to establish the relative similarity of these measures along with key properties and performance characteristics. While many measures are highly correlated, those commonly used within microbial ecology were found to be distinct from those popular within classical ecology, and from the recently recommended Gower and Canberra measures. Many of the measures are surprisingly robust to different rootings of the gene tree, the choice of similarity threshold used to define operational taxonomic units, and the presence of outlying basal lineages. Measures differ considerably in their sensitivity to rare organisms, and the effectiveness of measures can vary substantially under alternative models of differentiation. Consequently, the depth of sequencing required to reveal underlying patterns of relationships between environmental samples depends on the selected measure. Our results demonstrate that using complementary measures of phylogenetic β diversity can further our understanding of how communities are phylogenetically differentiated. Open-source software implementing the phylogenetic β-diversity measures evaluated in this manuscript is available at http://kiwi.cs.dal.ca/Software/ExpressBetaDiversity.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Barr JJ, Slater FR, Fukushima T, Bond PL . (2010). Evidence for bacteriophage activity causing community and performance changes in a phosphorus-removal activated sludge. FEMS Microbiol Ecol 74: 631–642.
Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL . (2008). Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci USA 105: 11505–11511.
Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J et al (2011). Moving pictures of the human microbiome. Genome Biol 12: R50.
Clarke KR, Warwick RM . (1998). A taxonomic distinctness index and its statistical properties. J Appl Ecol 35: 523–531.
Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A et al (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423.
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R . (2009). Bacterial community variation in human body habitats across space and time. Science 329: 1694–1697.
Crozier RH, Dunnett LJ, Agapow PM . (2005). Phylogenetic biodiversity assessment based on systematic nomenclature. Evol Bioninform Online 1: 11–36.
DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al (2006). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399.
Dethlefsen L, Huse S, Sogin ML, Relman DA . (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280.
Faith DP, Baker AM . (2007). Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online 17: 121–128.
Fierer N, Lauber CL, Zhou N, Daniel M, Costello EK, Knight R . (2010). Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA 107: 6477–6481.
Graham CH, Fine PVA . (2008). Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol Lett 11: 1265–1277.
Green JL, Plotkin JB . (2007). A statistical theory for sampling species abundances. Ecol Lett 10: 1037–1045.
Hamady M, Lozupone C, Knight R . (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27.
Horner-Devine MC, Bohannan BJM . (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: S100–S108.
Kluge AG, Farris JS . (1969). Quantitative phyletics and the evolution of anurans. Syst Zool 18: 1–32.
Koleff P, Gaston KJ, Lennon JJ . (2003). Measuring beta diversity for presence absence data. J Anim Ecol 72: 367–382.
Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R . (2010). Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods 7: 813–819.
Lauber CL, Hamady M, Knight R, Fierer N . (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community composition at the continental scale. Appl Environ Microbiol 75: 5111–5120.
Legendre P, Legendre L . (1998) Numerical Ecology 2nd English edn. Elsevier: Amsterdam, The Netherlands.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Lozupone CA, Hamady M, Kelley ST, Knight R . (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73: 1576–1585.
Lozupone CA, Hamady M, Knight R . (2006). UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440.
Magurran AE . (2004) Measuring Biological Diversity. Blackwell Publishing: Oxford, UK.
Martin AP . (2002). Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol 68: 3673–3682.
Martiny JB, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
Nipperess DA, Faith DP, Barton K . (2010). Resemblance in phylogenetic diversity among ecological assemblages. J Veg Sci 21: 809–820.
Pielou EC . (1984). The interpretation of ecological data: a primer of classification and ordination. John Wiley and Sons. Plotkin JB, Muller-Landau HC (eds). (2002). Sampling the species composition of a landscape. Ecology 83: 3344–3356.
Plotkin JB, Muller-Landau H . (2002). Sampling the species composition of a landscape. Ecology 83: 3344–3356.
Price MN, Dehal PS, Arkin AP . (2009). FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650.
Root HT, Nelson PR . (2011). Does phylogenetic distance aid in detecting environmental gradients related to species composition? J Veg Sci 22: 1143–1148.
Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG et al (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4: 1340–1351.
Schloss PD . (2008). Evaluating different approaches that test whether microbial communities have the same structure. ISME J 2: 265–275.
Schloss PD, Handelsman J . (2004). Status of the microbial census. Mol Biol Rev 68: 686–691.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Slater FR, Johnson CR, Blackall LL, Beiko RG, Bond PL . (2010). Monitoring associations between clade-level variation, overall community structure and ecosystem function in enhanced biological phosphorus removal (EBPR) systems using terminal-restriction fragment length polymorphism (T-RFLP). Water Res 44: 4908–4923.
Stackebrandt E, Goebel BM . (1994). Taxonomic Note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44: 846–849.
Swenson NG . (2011). Phylogenetic beta diversity metrics, trait evolution and inferring the functional beta diversity of communities. PLOS One 6: e21264.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484.
Ubeda C, Taur Y, Jeng RR, Equinda MJ, Son T, Samstein M et al (2010). Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120: 4332–4341.
Webb CO, Ackerly DD, Kembel SW . (2008). Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24: 2098–2100.
Acknowledgements
DHP was supported by the Killam Trusts; RGB acknowledges the support of Genome Atlantic, the Canada Foundation for Innovation, and the Canada Research Chairs program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Parks, D., Beiko, R. Measures of phylogenetic differentiation provide robust and complementary insights into microbial communities. ISME J 7, 173–183 (2013). https://doi.org/10.1038/ismej.2012.88
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.88
Keywords
This article is cited by
-
New insights from the biogas microbiome by comprehensive genome-resolved metagenomics of nearly 1600 species originating from multiple anaerobic digesters
Biotechnology for Biofuels (2020)
-
Using null models to compare bacterial and microeukaryotic metacommunity assembly under shifting environmental conditions
Scientific Reports (2020)
-
Actinobacteria from Antarctica as a source for anticancer discovery
Scientific Reports (2020)
-
HIV-associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases
Nature Communications (2020)
-
Distinct Profiles in Microbial Diversity on Carbon Steel and Different Welds in Simulated Marine Microcosm
Current Microbiology (2020)