Abstract
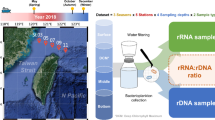
Very little is known about growth rates of individual bacterial taxa and how they respond to environmental flux. Here, we characterized bacterial community diversity, structure and the relative abundance of 16S rRNA and 16S rRNA genes (rDNA) using pyrosequencing along the salinity gradient in the Delaware Bay. Indices of diversity, evenness, structure and growth rates of the surface bacterial community significantly varied along the transect, reflecting active mixing between the freshwater and marine ends of the estuary. There was no positive correlation between relative abundances of 16S rRNA and rDNA for the entire bacterial community, suggesting that abundance of bacteria does not necessarily reflect potential growth rate or activity. However, for almost half of the individual taxa, 16S rRNA positively correlated with rDNA, suggesting that activity did follow abundance in these cases. The positive relationship between 16S rRNA and rDNA was less in the whole water community than for free-living taxa, indicating that the two communities differed in activity. The 16S rRNA:rDNA ratios of some typically marine taxa reflected differences in light, nutrient concentrations and other environmental factors along the estuarine gradient. The ratios of individual freshwater taxa declined as salinity increased, whereas the 16S rRNA:rDNA ratios of only some typical marine bacteria increased as salinity increased. These data suggest that physical and other bottom-up factors differentially affect growth rates, but not necessarily abundance of individual taxa in this highly variable environment.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Andersson AF, Riemann L, Bertilsson S . (2010). Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J 4: 171–181.
Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF . (2001). Proteorhodopsin phototrophy in the ocean. Nature 411: 786–789.
Bouvier TC, del Giorgio PA . (2002). Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr 47: 453–470.
Bremer H, Dennis PP . (1996). Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al. (eds). Escherichia coli and Salmonella: Cellular And Molecular Biology. ASM Press: Washington, DC, pp 1553–1569.
Buchan A, Gonzalez JM, Moran MA . (2005). Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71: 5665–5677.
Campbell BJ, Yu L, Heidelberg JF, Kirchman DL . (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781.
Campbell BJ, Yu L, Straza TRA, Kirchman DL . (2009). Temporal changes in bacterial rRNA and rRNA genes in Delaware (USA) coastal waters. Aquat Microb Ecol 57: 123–135.
Church TM . (1986). Biogeochemical factors influencing the residence time of microconstituents in a large tidal estuary, Delaware Bay. Mar Chem 18: 393–406.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Cottrell MT, Kirchman DL . (2004). Single-cell analysis of bacterial growth, cell size, and community structure in the Delaware estuary. Aquat Microb Ecol 34: 139–149.
Cottrell MT, Mannino A, Kirchman DL . (2006). Aerobic anoxygenic phototrophic bacteria in the Mid-Atlantic Bight and the North Pacific Gyre. Appl Environ Microbiol 72: 557–564.
Crump BC, Baross JA . (1996). Particle-attached bacteria and heterotrophic plankton associated with the Columbia River estuarine turbidity maxima. Mar Ecol Progr Ser 138: 265–273.
Crump BC, Baross JA . (2000). Characterization of the bacterially-active particle fraction in the Columbia River estuary. Mar Ecol Progr Ser 206: 13–22.
Crump BC, Baross JA, Simenstad CA . (1998). Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol 14: 7–18.
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE . (2004). Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70: 1494–1505.
Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ . (1999). Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques 27: 66–68.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Deutscher MP . (2006). Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 34: 659–666.
Ducklow HW . (2000). Bacterial production and biomass in the oceans. In: Kirchman DL, (ed). Microbial Ecology of the Oceans. Wiley: New York, pp 85–120.
Fegatella F, Lim J, Kjelleberg S, Cavicchioli R . (1998). Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol 64: 4433–4438.
Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC . (2012). Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J 6: 554–563.
Fouilland E, Mostajir B . (2010). Revisited phytoplanktonic carbon dependency of heterotrophic bacteria in freshwaters, transitional, coastal and oceanic waters. FEMS Microbiol Ecol 73: 419–429.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
Gaidos E, Rusch A, Ilardo M . (2011). Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol 13: 1138–1152.
Ghiglione JF, Mevel G, Pujo-Pay M, Mousseau L, Lebaron P, Goutx M . (2007). Diel and seasonal variations in abundance, activity, and community structure of particle-attached and free-living bacteria in NW Mediterranean Sea. Microbial Ecol 54: 217–231.
Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139.
Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL et al. (2005). Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Giovannoni SJ, Vergin KL . (2012). Seasonality in ocean microbial communities. Science 335: 671–676.
Goosen NK, Kromkamp J, Peene J, van Rijswik P, van Breugel P . (1999). Bacterial and phytoplankton production in the maximum turbidity zone of three European estuaries: the Elbe, Westerschelde and Gironde. J Mar Syst 22: 151–171.
Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL et al. (2010). Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol 8: e1000358.
Hammer O, Harper DAT, Ryan PD . (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4: 9.
Heidelberg JF, Heidelberg KB, Colwell RR . (2002). Seasonality of Chesapeake Bay bacterioplankton species. Appl Environ Microbiol 68: 5488–5497.
Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF . (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5: 1571–1579.
Hoch MP, Kirchman DL . (1993). Seasonal and interannual variability in bacterial production and biomass in a temperate estuary. Mar Ecol Prog Ser 98: 283–295.
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL . (2008). NCBI BLAST: a better web interface. Nucleic Acids Res 36: W5–W9.
Jones SE, Lennon JT . (2010). Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA 107: 5881–5886.
Kan J, Evans SE, Chen F, Suzuki MT . (2008). Novel estuarine bacterioplankton in rRNA operon libraries from the Chesapeake Bay. Aquat Microb Ecol 51: 55–66.
Kan JJ, Crump BC, Wang K, Chen F . (2006). Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol Oceanogr 51: 2157–2169.
Kemp PF, Lee S, LaRoche J . (1993). Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol 59: 2594–2601.
Kerkhof L, Kemp P . (1999). Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol 30: 253–260.
Kirchman DL . (1993). Particulate detritus and bacteria in marine environments. In: Ford T, (ed). Aquatic Microbiology: An Ecological Approach. Blackwell: Cambridge, MA, pp 321–341.
Kirchman DL, Dittel AI, Malmstrom RR, Cottrell MT . (2005). Biogeography of major bacterial groups in the Delaware estuary. Limnol Oceanogr 50: 1697–1706.
Kirchman DL, Yu LY, Fuchs BM, Amann R . (2001). Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat Microb Ecol 26: 13–22.
Kramer JG, Singleton FL . (1992). Variations in rRNA content of marine Vibrio spp. during starvation–survival and recovery. Appl Environ Microbiol 58: 201–207.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440.
Michelou VK, Cottrell MT, Kirchman DL . (2007). Light-stimulated bacterial production and amino acid assimilation by cyanobacteria and other microbes in the North Atlantic Ocean. Appl Environ Microbiol 73: 5539–5546.
Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ . (2005). Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50: 1687–1696.
Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144.
Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S . (2011). A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75: 14–49.
Nilsson M, Bulow L, Wahlund KG . (1997). Use of flow field-flow fractionation for the rapid quantitation of ribosome and ribosomal subunits in Escherichia coli at different protein production conditions. Biotechnol Bioeng 54: 461–467.
Preen K, Kirchman DL . (2004). Microbial respiration and production in the Delaware Estuary. Aquat Microb Ecol 37: 109–119.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Puddu A, La Ferla R, Allegra A, Bacci C, Lopez M, Oliva F et al. (1998). Seasonal and spatial distribution of bacterial production and biomass along a salinity gradient (Northern Adriatic Sea). Hydrobiologia 363: 271–282.
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ . (2011). Removing noise from pyrosequenced amplicons. BMC Bioinform 12: 38.
Rappé MS, Connon SA, Vergin KL, Giovannoni SJ . (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: 398–431.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Sharp JH, Yoshiyama K, Parker AE, Schwartz MC, Curless SE, Beauregard AY et al. (2009). A biogeochemical view of estuarine eutrophication: seasonal and spatial trends and correlations in the Delaware Estuary. Estuar Coast 32: 1023–1043.
Šimek K, Horňàk K, Jezbera J, Nedoma J, Vrba J, Stralkrábová V et al. (2006). Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ Microbiol 8: 1613–1624.
Simon M, Grossart HP, Schweitzer B, Ploug H . (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol 28: 175–211.
Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ . (2011). Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One 6: e19725.
Straza TRA, Kirchman DL . (2011). Single-cell response of bacterial groups to light and other environmental factors in the Delaware Bay, USA. Aquat Microb Ecol 62: 267–U281.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739.
Telesh IV, Khlebovich VV . (2010). Principal processes within the estuarine salinity gradient: a review. Mar Pollut Bull 61: 149–155.
Yokokawa T, Nagata T, Cottrell MT, Kirchman DL . (2004). Growth rate of the major phylogenetic bacterial groups in the Delaware estuary. Limnol Oceanogr 49: 1620–1629.
Acknowledgements
We thank Matt Cottrell and Sharon Grim for sampling assistance and Liying Yu for technical assistance. This work was supported by National Science Foundation grants to BJC and DLK (OCE-0825468) and to DLK (MCB-0453993 and OCE-1030306).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Campbell, B., Kirchman, D. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7, 210–220 (2013). https://doi.org/10.1038/ismej.2012.93
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2012.93
Keywords
This article is cited by
-
Subsidy-stress responses of ecosystem functions along experimental freshwater salinity gradients
Biogeochemistry (2024)
-
Impact of environmental factors on diversity of fungi in sediments from the Shenzhen River Estuary
Archives of Microbiology (2023)
-
Abundant and Rare Taxa of Planktonic Fungal Community Exhibit Distinct Assembly Patterns Along Coastal Eutrophication Gradient
Microbial Ecology (2023)
-
Coral-associated nitrogen fixation rates and diazotrophic diversity on a nutrient-replete equatorial reef
The ISME Journal (2022)
-
Soil bacterial and fungal communities respond differently to Bombax ceiba (Malvaceae) during reproductive stages of rice in a traditional agroforestry system
Plant and Soil (2022)