Abstract
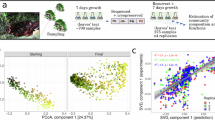
Microbial communities mediate crucial biogeochemical, biomedical and biotechnological processes, yet our understanding of their assembly, and our ability to control its outcome, remain poor. Existing evidence presents conflicting views on whether microbial ecosystem assembly is predictable, or inherently unpredictable. We address this issue using a well-controlled laboratory model system, in which source microbial communities colonize a pristine environment to form complex, nutrient-cycling ecosystems. When the source communities colonize a novel environment, final community composition and function (as measured by redox potential) are unpredictable, although a signature of the community’s previous history is maintained. However, when the source communities are pre-conditioned to their new habitat, community development is more reproducible. This situation contrasts with some studies of communities of macro-organisms, where strong selection under novel environmental conditions leads to reproducible community structure, whereas communities under weaker selection show more variability. Our results suggest that the microbial rare biosphere may have an important role in the predictability of microbial community development, and that pre-conditioning may help to reduce unpredictability in the design of microbial communities for biotechnological applications.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Anderson MJ, Gorely RN, Clarke KR . (2008) PERMANOVA+ PRIMER: Guide to Software and Statistical Methods. Primer-E: Plymouth, UK.
Ayarza JM, Erijman L . (2011). Balance of neutral and deterministic components in the dynamics of activated sludge floc assembly. Microb Ecol 61: 486–495.
Baptista J de C, Davenport RJ, Donnelly T, Curtis TP . (2008). The microbial diversity of laboratory-scale wetlands appears to be randomly assembled. Water Res 42: 3182–3190.
Becks L, Hilker FM, Malchow H, Jürgens K, Arndt H . (2005). Experimental demonstration of chaos in a microbial food web. Nature 435: 1226–1229.
Benincà E, Huisman J, Heerkloss R, Jöhnk KD, Branco P, Van Nes EH et al (2008). Chaos in a long-term experiment with a plankton community. Nature 451: 822–825.
Black AJ, McKane AJ . (2012). Stochastic formulation of ecological models and their applications. Trends Ecol Evol 27: 337–345.
Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS . (2012). Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl Environ Microbiol 78: 3221–3228.
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T . (2011). Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108: 14288–14293.
Caruso T, Chan Y, Lacap DC, Lau MCY, McKay CP, Pointing SB . (2011). Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J 5: 1406–1413.
Chase JM . (2003). Community assembly: when should history matter? Oecologia 136: 489–498.
Chase JM . (2007). Drought mediates the importance of stochastic community assembly. Proc Natl Acad Sci USA 104: 17430–17434.
Clarke KR, Warwick RM . (2001) Change in Marine Communities: An Approach to Statistical Analysis. 2nd edn Primer-E: Plymouth, UK.
Donohue I, Jackson AL, Pusch MT, Irvine K . (2009). Nutrient enrichment homogenizes lake benthic assemblages at local and regional scales. Ecology 90: 3470–3477.
Fernández A, Huang S, Seston S, Xing J, Hickey R, Criddle C et al (1999). How stable is stable? Function versus community composition. Appl Environ Microbiol 65: 3697–3704.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
Geets J, Borremans B, Diels L, Springael D, Vangronsveld J, van der Lelie D et al (2006). DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J Microbiol Methods 66: 194–205.
Graham DW, Knapp CW, Van Vleck ES, Bloor K, Lane TB, Graham CE . (2007). Experimental demonstration of chaotic instability in biological nitrification. ISME J 1: 385–393.
Hekstra DR, Leibler S . (2012). Contingency and statistical laws in replicate microbial closed ecosystems. Cell 149: 1164–1173.
Helmus MR, Keller W, Paterson MJ, Yan ND, Cannon CH, Rusak JA . (2010). Communities contain closely related species during ecosystem disturbance. Ecol Letts 13: 162–174.
Jessup CM, Kassen R, Forde SE, Kerr B, Buckling A, Rainey PB et al (2004). Big questions, small worlds: microbial model systems in ecology. Trends Ecol Evol 19: 189–197.
Jiang L, Patel SN . (2008). Community assembly in the presence of disturbance: A microcosm experiment. Ecology 89: 1931–1940.
Khatri BS, Free A, Allen RJ . (2012). Oscillating microbial dynamics driven by small populations, limited nutrient supply and high death rates. J Theor Biol 314: 120–129.
Knope ML, Forde SE, Fukami T . (2012). Evolutionary history, immigration history, and the extent of diversification in community assembly. Front Microbiol 2: article 273.
Kruskal JB . (1964). Multidimensional-scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29: 1–27.
Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R . (2010). Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods 7: 813–819.
Kurtz JC, Devereu R, Barkay T, Jonas RB . (1998). Evaluation of sediment slurry microcosms for modeling microbial communities in estuarine sediments. Environ Toxicol Chem 17: 1274–1281.
Langenheder S, Székely AJ . (2011). Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5: 1086–1094.
Langenheder S, Lindstrom ES, Tranvik LJ . (2006). Structure and function of bacterial communities emerging from different sources under identical conditions. Appl Environ Microbiol 72: 212–220.
Lazzaro A, Gauer A, Zeyer J . (2011). Field-scale transplantation experiment to investigate structures of soil bacterial communities at pioneering sites. Appl Environ Microbiol 77: 8241–8248.
Lepori F, Malmqvist B . (2009). Deterministic control on community assembly peaks at intermediate levels of disturbance. Oikos 118: 471–479.
Madigan RT, Martinko J, Stahl DA, Clark DP . (2011) Brock Biology of Microorganisms 13th edn. Pearson Prentice Hall: London.
Martiny JBH, Bohannan BJM, Borwn JH, Colwell RK, Fuhrman JA, Green JL et al (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
McCaig AE, Glover LA, Prosser JI . (2001). Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl Environ Microbiol 67: 4554–4559.
Muyzer G, de Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl Environ Microbiol 59: 695–700.
Myers JA, Harms KE . (2011). Seed arrival and ecological filters interact to assemble high-diversity plant communities. Ecology 92: 676–686.
Nicol GW, Tscherko D, Embley TM, Prosser JI . (2005). Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol 7: 337–347.
Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA et al (2010). Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA 107: 15345–15350.
Padua RA, Parrado A, Larghero J, Chomienne C . (1999). UV and clean air result in contamination-free PCR. Leukemia 13: 1898–1899.
Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO . (2007). Development of the human infant intestinal microbiota. PLoS Biol 5: 1556–1573.
Park H, Rosenthal A, Jezek R, Ramalingam K, Fillos J, Chandran K . (2010). Impact of inocula and growth mode on the molecular microbial ecology of anaerobic ammonia oxidation (anammox) bioreactor communities. Water Res 44: 5005–5013.
Pedrós-Alió C . (2012). The rare bacterial biosphere. Ann Rev Marine Sci 4: 449–466.
Prosser JI . (2012). Ecosystem processes and interactions in a morass of diversity. FEMS Microbiol Ecol 81: 507–519.
Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP et al (2007). The role of ecological theory in microbial ecology. Nat Rev Microbiol 5: 384–392.
Roeselers G, Zippel B, Staal M, van Loosdrecht M, Muyzer G . (2006). On the reproducibility of microcosm experiments - different community composition in parallel phototrophic biofilm microcosms. FEMS Microbiol Ecol 58: 169–178.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Sokal RR, Rohlf FJ . (1969) Biometry: The Principles and Practice of Statistics in Biological Research. W.H. Freeman and Company: San Francisco.
Trosvik P, Stenseth NC, Rudi K . (2010). Convergent temporal dynamics of the human infant gut microbiota. ISME J 4: 151–158.
Violle C, Pu ZC, Jiang L . (2010). Experimental demonstration of the importance of competition under disturbance. Proc Natl Acad Sci USA 107: 12925–12929.
Weatherby AJ, Warren PH, Law R . (1998). Coexistence and collapse: an experimental investigation of the persistent communities of a protist species pool. J Animal Ecol 67: 554–566.
Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Guillaumaud N et al (2007). Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ Microbiol 9: 2211–2219.
Whitman WB, Coleman DC, Wiebe WJ . (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583.
Acknowledgements
The authors thank Richard Blythe, Tom Curtis, Neil Gray, Bhavin Khatri, Jana Schwarz-Linek, Wilson Poon, Jim Prosser and Bartlomiej Waclaw for valuable discussions, Kate Heal, Bhavin Khatri and Alex Jackson for assistance with sample collection, Martin Simmen for assistance with NMDS and statistical analysis, Jan Krowkowski for environmental data and Stephen Bridgett and Chris Quince for assistance with the high-throughput sequence analysis. BMS was supported through WP 6 of the EU FP7 WISER project. MEC holds a Royal Society Research Professorship, while RJA was supported by the Royal Society of Edinburgh and by a Royal Society University Research Fellowship. AF was supported by a Darwin Trust Fellowship, and thanks the late Prof. Kenneth Murray for additional support. This work was funded by the Leverhulme Trust under grant F/00158/BX, by the EPSRC under grant EP/E030173, by a NERC CEH small grant and by a Royal Society Research Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Pagaling, E., Strathdee, F., Spears, B. et al. Community history affects the predictability of microbial ecosystem development. ISME J 8, 19–30 (2014). https://doi.org/10.1038/ismej.2013.150
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.150
Keywords
This article is cited by
-
Functional conservation of microbial communities determines composition predictability in anaerobic digestion
The ISME Journal (2023)
-
Soil pH indirectly determines Ralstonia solanacearum colonization through its impacts on microbial networks and specific microbial groups
Plant and Soil (2023)
-
Rare soil bacteria are more responsive in desertification restoration than abundant bacteria
Environmental Science and Pollution Research (2022)
-
Environmental filtering of bacterial functional diversity along an aridity gradient
Scientific Reports (2019)
-
Historical contingency and productivity effects on food-chain length
Communications Biology (2019)