Abstract
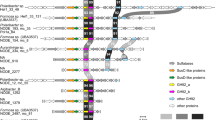
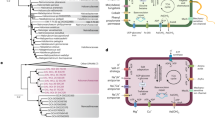
We investigated the mechanisms of osmoadaptation in the order Halobacteriales, with special emphasis on Haladaptatus paucihalophilus, known for its ability to survive in low salinities. H. paucihalophilus genome contained genes for trehalose synthesis (trehalose-6-phosphate synthase/trehalose-6-phosphatase (OtsAB pathway) and trehalose glycosyl-transferring synthase pathway), as well as for glycine betaine uptake (BCCT family of secondary transporters and QAT family of ABC transporters). H. paucihalophilus cells synthesized and accumulated ∼1.97–3.72 μmol per mg protein of trehalose in a defined medium, with its levels decreasing with increasing salinities. When exogenously supplied, glycine betaine accumulated intracellularly with its levels increasing at higher salinities. RT-PCR analysis strongly suggested that H. paucihalophilus utilizes the OtsAB pathway for trehalose synthesis. Out of 83 Halobacteriales genomes publicly available, genes encoding the OtsAB pathway and glycine betaine BCCT family transporters were identified in 38 and 60 genomes, respectively. Trehalose (or its sulfonated derivative) production and glycine betaine uptake, or lack thereof, were experimentally verified in 17 different Halobacteriales species. Phylogenetic analysis suggested that trehalose synthesis is an ancestral trait within the Halobacteriales, with its absence in specific lineages reflecting the occurrence of gene loss events during Halobacteriales evolution. Analysis of multiple culture-independent survey data sets demonstrated the preference of trehalose-producing genera to saline and low salinity habitats, and the dominance of genera lacking trehalose production capabilities in permanently hypersaline habitats. This study demonstrates that, contrary to current assumptions, compatible solutes production and uptake represent a common mechanism of osmoadaptation within the Halobacteriales.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G . (2006). Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6: 109.
Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E . (2008). Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 12: 505–518.
Bidle K, Amadio W, Oliveira P, Paulish T, Sharron H, Earnest C . (2005). A phylogenetic analysis of Haloarchaea found in a solar saltern. Bios 76: 89–96.
Bodaker I, Sharon I, Suzuki MT, Feingersch R, Shmoish M, Andreishcheva E et al. (2009). Comparative community genomics in the Dead Sea: an increasingly extreme environment. ISME J 4: 399–407.
Bowman JP, McCammon SA, Rea SM, McMeekin TA . (2000). The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett 183: 81–88.
Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML . (2004). Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most Haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl Environ Microbiol 70: 5258–5265.
Christian JH, Waltho JA . (1962). Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim Biophys Acta 65: 506–508.
Desmarais D, Jablonski PE, Fedarko NS, Roberts MF . (1997). 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J Bacteriol 179: 3146–3153.
Ellis DG, Bizzoco RW, Kelley ST . (2008). Halophilic Archaea determined from geothermal steam vent aerosols. Environ Microbiol 10: 1582–1590.
Elshahed MS, Najar FZ, Roe BA, Oren A, Dewers TA, Krumholz LR . (2004). Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl Environ Microbiol 70: 2230–2239.
Enache M, Takashi I, Fukushima T, Usami R, Dumitru L, Kamekura M . (2007). Phylogenetic relationships within the family Halobacteriaceae inferred from rpoB’ gene and protein sequences. Int J Syst Evol Microbiol 57: 2289–2295.
Ginzburg M, Sachs L, Ginzburg BZ . (1970). Ion metabolism in a Halobacterium: I. Influence of age of culture on intracellular concentrations. J Gen Physiol 55: 187–207.
Goh F, Jeon YJ, Barrow K, Neilan BA, Burns BP . (2011). Osmoadaptive strategies of the archaeon Halococcus hamelinensis Isolated from a hypersaline stromatolite environment. Astrobiology 11: 529–536.
Grant WD . (2004). Life at low water activity. Philos Trans Roy Soc B 359: 1249–1267.
Grieve CM, Grattan SR . (1983). Rapid assay for determination of water soluble quaternary ammonium compounds. Plant Soil 70: 303–307.
Hagemann M . (2011). Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35: 87–123.
Inoue K, Itoh T, Ohkuma M, Kogure K . (2011). Halomarina oriensis gen. nov., sp. nov., a halophilic archaeon isolated from a seawater aquarium. Int J Syst Evol Microbiol 61: 942–846.
Kokoeva MV, Storch KF, Klein C, Oesterhelt D . (2002). A novel mode of sensory transduction in archaea: binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J 15: 2312–2322.
Lanyi JK, Silverman MP . (1972). The state of binding of intracellular K+in Halobacterium cutirubrum. Can J Microbiol 18: 993–995.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948.
Lundquist P, Mårtensson J, Sörbo B, Ohman S . (1980). Turbidimetry of inorganic sulfate, ester sulfate, and total sulfur in urine. Clin Chem 26: 1178–1181.
Mani K, Salgaonkar B, Braganca J . (2012). Culturable halophilic archaea at the initial and crystallization stages of salt production in a natural solar saltern of Goa, India. Aquat Biosyst 8: 15.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acid Res 40: D115–D122.
Matheson AT, Sprott GD, McDonald IJ, Tessier H . (1976). Some properties of an unidentified halophile: growth characteristics, internal salt concentration, and morphology. Can J Microbiol 22: 780–786.
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T . (2010). Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B' (rpoB') gene. Int J Syst Evol Microbiol 60: 2398–2408.
Mojica FJ, Cisneros E, Ferrer C, Rodríguez-Valera F, Juez G . (1997). Osmotically induced response in representatives of halophilic prokaryotes: the bacterium Halomonas elongata and the archaeon Haloferax volcanii. J Bacteriol 179: 5471–5481.
Mougous JD, Petzold CJ, Senaratne RH, Lee DH, Akey DL, Lin FL et al. (2004). Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nat Struct Mol Biol 11: 721–729.
Munson MA, Nedwell DB, Embley TM . (1997). Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol 63: 4729–4733.
Nicolaus B, Gambacorta A, Basso AL, Riccio R, Rosa MD, Grant WD . (1988). Trehalose in Archaebacteria. Syst Appl Microbiol 10: 215–217.
Oh D, Porter K, Russ B, Burns D, Dyall-Smith M . (2010). Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14: 161–169.
Oren A . (1999). Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev 63: 334–348.
Oren A . (2005). A hundred years of Dunaliella research: 1905–2005. Saline Syst 1: 2.
Oren A . (2010). Mats of filamentous and unicellular Cyanobacteria in hypersaline environments. In: Seckbach J, Oren A (eds). Microbial Mats. Springer: Netherlands, pp 387–400.
Oren A . (2013). Life at high salt concentrations. The Prokaryotes—Prokaryotic Communities and Ecophysiology (4th edn). Springer: Berlin-Heidelburg.
Oren A, Ginzburg M, Ginzburg BZ, Hochstein LI, Volcani BE . (1990). Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Intl J Syst Bacteriol 40: 209–210.
Oren A, Heldal M, Norland S, Galinski E . (2002a). Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles 6: 491–498.
Oren A, Mana L . (2002b). Amino acid composition of bulk protein and salt relationships of selected enzymes of Salinibacter ruber, an extremely halophilic bacterium. Extremophiles 6: 217–223.
Perreault NN, Andersen DT, Pollard WH, Greer CW, Whyte LG . (2007). Characterization of the prokaryotic diversity in cold saline perennial springs of the Canadian high arctic. Appl Environ Microbiol 73: 1532–1543.
Plackett RL . (1983). Karl Pearson and the chi-squared test. Int Statist Rev 51: 59–72.
Podell S, Ugalde JA, Narasingarao P, Banfield JF, Heidelberg KB, Allen EE . (2013). Assembly-driven community genomics of a hypersaline microbial ecosystem. PLoS One 8: e61692.
Purdy KJ, Cresswell-Maynard TD, Nedwell DB, McGenity TJ, Grant WD, Timmis KN et al. (2004). Isolation of haloarchaea that grow at low salinities. Environ Microbiol 6: 591–595.
Pérez-Fillol M, Rodríguez-Valera F . (1986). Potassium ion accumulation in cells of different halobacteria. Microbiologia (Madrid, Spain) 2: 73–80.
R Development Core Team (2011) R: A Language and Environment for Statistical Computing. Reference Index., 2.13 (2nd edn) R Foundation for Statistical Computing: Vienna, Austria.
Regeva R, Peria I, Gilboa H, Avi-Dor Y . (1990). 13C NMR study of the interrelation between synthesis and uptake of compatible solutes in two moderately halophilic eubacteria: Bacterium Ba1 and Vibro costicola. Arch Biochem Biophys 278: 106–112.
Roberts M . (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1: 5.
Savage KN, Krumholz LR, Oren A, Elshahed MS . (2007). Haladaptatus paucihalophilus gen. nov., sp. nov., a halophilic archaeon isolated from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol 57: 19–24.
Stivaletta N, Barbieri R, Cevenini F, López-García P . (2011). Physicochemical conditions and microbial diversity associated with the evaporite deposits in the Laguna de la Piedra (Salar de Atacama, Chile). Geomicrobiol J 28: 83–95.
Takagi M, Nakamura H, Sanui Y, Ueno K . (1981). Spectrophotometric determination of sodium by ion-pair extraction with crown ether complexes and monoanionic dyes. Analyt Chim Acta 126: 185–190.
Takai K, Komatsu T, Inagaki F, Horikoshi F . (2001). Distribution of archaea in a black smoker chimney structure. Appl Environ Microbiol 67: 3618–3629.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739.
Uzawa H, Nishida Y, Sasaki K, Nagatsuka T, Hiramatsu H, Kobayashi K . (2004). Sulfatase-catalyzed assembly of regioselectively O-sulfonated p-nitrophenyl α-d-gluco- and α-d-mannopyranosides. Carbohyd Res 339: 1597–1602.
Ventosa A, Oren A . (1996). Halobacterium salinarum nom. corrig., a name to replace Halobacterium salinarium (Elazari-Volcani) and to include Halobacterium halobium and Halobacterium cutirubrum. Int J Syst Bacteriol 46: 347–347.
Ventosa A, Rodriguez-Valera F, Poindexter JS, Reznikoff WS . (1984). Selection for moderately halophilic bacteria by gradual salinity increases. Can J Microbiol 30: 1279–1282.
Walsh DA, Bapteste E, Kamekura M, Doolittle WF . (2004). Evolution of the RNA polymerase B′ subunit gene (rpoB′) in Halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol Biol Evol 21: 2340–2351.
Walsh DA, Papke RT, Doolittle WF . (2005). Archaeal diversity along a soil salinity gradient prone to disturbance. Environ Microbiol 7: 1655–1666.
Xiao W, Wang Z-G, Wang Y-X, Schneegurt MA, Li Z-Y, Lai Y-H et al. (2013). Comparative molecular analysis of the prokaryotic diversity of two salt mine soils in southwest China. J Basic Microbiol doi:10.1002/jobm.201200200.
Youssef NH, Ashlock-Savage KN, Elshahed MS . (2012). Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl Environ Microbiol 78: 1332–1344.
Acknowledgements
We thank Ratnakar Deole and Audra Liggenstoffer for technical assistance, Margret Eastman for NMR analysis and Ramy Aziz for helpful discussions. This work was supported by the National Science Foundation Microbial Observatories Program (grant EF0801858).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Youssef, N., Savage-Ashlock, K., McCully, A. et al. Trehalose/2-sulfotrehalose biosynthesis and glycine-betaine uptake are widely spread mechanisms for osmoadaptation in the Halobacteriales. ISME J 8, 636–649 (2014). https://doi.org/10.1038/ismej.2013.165
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.165
Keywords
This article is cited by
-
Assessment of diversity of archaeal communities in Algerian chott
Extremophiles (2023)
-
Physiological and genomic insights into abiotic stress of halophilic archaeon Natrinema altunense 4.1R isolated from a saline ecosystem of Tunisian desert
Genetica (2023)
-
Hly176B, a low-salt tolerant halolysin from the haloarchaeon Haloarchaeobius sp. FL176
World Journal of Microbiology and Biotechnology (2023)
-
Revealing the salinity adaptation mechanism in halotolerant bacterium Egicoccus halophilus EGI 80432T by physiological analysis and comparative transcriptomics
Applied Microbiology and Biotechnology (2021)
-
Current developments on polyhydroxyalkanoates synthesis by using halophiles as a promising cell factory
Microbial Cell Factories (2020)