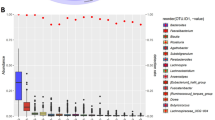
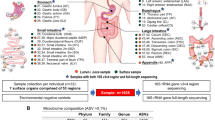
Abstract
Human gut microbiota shows high inter-subject variations, but the actual spatial distribution and co-occurrence patterns of gut mucosa microbiota that occur within a healthy human instestinal tract remain poorly understood. In this study, we illustrated a model of this mucosa bacterial communities’ biogeography, based on the largest data set so far, obtained via 454-pyrosequencing of bacterial 16S rDNAs associated with 77 matched biopsy tissue samples taken from terminal ileum, ileocecal valve, ascending colon, transverse colon, descending colon, sigmoid colon and rectum of 11 healthy adult subjects. Borrowing from macro-ecology, we used both Taylor’s power law analysis and phylogeny-based beta-diversity metrics to uncover a highly heterogeneous distribution pattern of mucosa microbial inhabitants along the length of the intestinal tract. We then developed a spatial dispersion model with an R-squared value greater than 0.950 to map out the gut mucosa-associated flora’s non-linear spatial distribution pattern for 51.60% of the 188 most abundant gut bacterial species. Furthermore, spatial co-occurring network analysis of mucosa microbial inhabitants together with occupancy (that is habitat generalists, specialists and opportunist) analyses implies that ecological relationships (both oppositional and symbiotic) between mucosa microbial inhabitants may be important contributors to the observed spatial heterogeneity of mucosa microbiota along the human intestine and may even potentially be associated with mutual cooperation within and functional stability of the gut ecosystem.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180.
Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M . (2008). Computing topological parameters of biological networks. Bioinformatics 24: 282–284.
Barberan A, Bates ST, Casamayor EO, Fierer N . (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343–351.
Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R et al (2007). Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J 1: 403–418.
Bown RL, Gibson JA, Sladen GE, Hicks B, Dawson AM . (1974). Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut 15: 999–1004.
Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E et al (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 108: 4586–4591.
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R . (2009). Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697.
Croix JA, Carbonero F, Nava GM, Russell M, Greenberg E, Gaskins HR . (2011). On the relationship between sialomucin and sulfomucin expression and hydrogenotrophic microbes in the human colonic mucosa. PLoS One 6: e24447.
Cuív PÓ, Klaassens ES, Durkin AS, Harkins DM, Foster L, McCorrison J et al (2011). Draft genome sequence of Bacteroides vulgatus PC510, a strain isolated from human feces. J Bacteriol 193: 4025–4026.
Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT . (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28: 1221–1227.
de Carcer DA, Cuiv PO, Wang T, Kang S, Worthley D, Whitehall V et al (2011). Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J 5: 801–809.
Drossel B, McKane AJ, Quince C . (2004). The impact of nonlinear functional responses on the long-term evolution of food web structure. J Theor Biol 229: 539–548.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M et al (2005). Diversity of the human intestinal microbial flora. Science 308: 1635–1638.
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J et al (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8: e1002606.
Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR . (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785.
Good IJ . (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264.
Girvan M, Newman MEJ . (2002). Community structure in social and biological networks. Proc Natl Acad Sci USA 99: 7821–7826.
Hayashi H, Sakamoto M, Benno Y . (2002). Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol 46: 535–548.
Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y . (2005). Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol 54: 1093–1101.
Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ . (2002). Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol 39: 33–39.
Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI . (2011). Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One 6: e25042.
Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P et al (2011). Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60: 631–637.
Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI . (2011). Human nutrition, the gut microbiome and the immune system. Nature 474: 327–336.
Kemp PF, Aller JY . (2004). Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol 47: 161–177.
Kilpatrick AM, Ives AR . (2003). Species interactions can explain Taylor's power law for ecological time series. Nature 422: 65–68.
Koropatkin NM, Cameron EA, Martens EC . (2012). How glycan metabolism shapes the human gut microbiota. Nat Rev Micro 10: 323–335.
Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A et al (2012). Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61: 1124–1131.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Lozupone C, Hamady M, Knight R . (2006). UniFrac - An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371.
Ma ZS . (2012). http://adsabs.harvard.edu/abs/2012arXiv1205.3504M.
Macfarlane GT, Gibson GR, Cummings JH . (1992). Comparison of fermentation reactions in different regions of the human colon. J Appl Microbiol 72: 57–64.
Moore WE, Holdeman LV . (1974). Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol 27: 961–979.
Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR . (2012). Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J 6: 57–70.
Newman MEJ . (2006). Modularity and community structure in networks. Proc Natl Acad Sci USA 103: 8577–8582.
Ochman H, Worobey M, Kuo C-H, Ndjango J-BN, Peeters M, Hahn BH et al (2010). Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65.
Rigottier-Gois L . (2013). Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7: 1256–1261.
Rivera C, Vakil R, Bader J . (2010). NeMo: network module identification in Cytoscape. BMC Bioinformatics 11: S61.
Rives AW, Galitski T . (2003). Modular organization of cellular networks. Proc Natl Acad Sci USA 100: 1128–1133.
Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A et al (2013). Genomic variation landscape of the human gut microbiome. Nature 493: 45–50.
Schluter J, Foster KR . (2012). The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol 10: e1001424.
Sheridan W, Lowndes R, Young H . (1987). Tissue oxygen tension as a predictor of colonic anastomotic healing. Dis Colon Rectum 30: 867–871.
Shigesada N, Kawasaki K, Teramoto E . (1979). Spatial segregation of interacting species. J Theor Biol 79: 83–99.
Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T . (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432.
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J et al (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736.
Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet J-P et al (2009). Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11: 2574–2584.
Taylor LR . (1961). Aggregation, variance and the mean. Nature 189: 732–735.
Taylor LR . (1984). Assessing and interpreting the spatial distributions of insect populations. Annu Rev Entomol 29: 321–357.
Taylor RAJ . (2007). Obituary: Roy (L. R.) Taylor (1924–2007). J Anim Ecol 76: 630–631.
Taylor LR, Taylor RAJ . (1977). Aggregation, migration and population mechanics. Nature 265: 415–421.
Taylor LR, Woiwod IP, Perry JN . (1979). The negative binomial as a dynamic ecological model for aggregation, and the density dependence of k. J Anim Ecol 48: 289–304.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484.
Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F et al (2010). Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA 107: 7503–7508.
Wang M, Ahrné S, Jeppsson B, Molin G . (2005). Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol 54: 219–231.
Wang X, Heazlewood SP, Krause DO, Florin THJ . (2003). Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol 95: 508–520.
Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L et al (2009). Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis 15: 653–660.
Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z et al (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139: 1844–1854.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M et al (2012). Human gut microbiome viewed across age and geography. Nature 486: 222–227.
Zhou Y, Gao H, Mihindukulasuriya K, Rosa P, Wylie K, Vishnivetskaya T et al (2013). Biogeography of the ecosystems of the healthy human body. Genome Biol 14: R1.
Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans ADL, de Vos WM . (2002). Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 68: 3401–3407.
Acknowledgements
The authors would like to thank Kunming Biological Diversity Regional Center of Large Apparatus and Equipments, Kunming Institute of Zoology, Chinese Academy of Sciences for their superb technical assistances as well as Andrew Willden (Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China) for improving the language of the manuscript. This research was supported by the following grants: National Natural Science Foundation of China (Grant No. 31100916 to ZZ and Grant No. 61175071 to ZM); Natural Science Foundation of Yunnan Province of China (Grant No. 2011FA035 to PS and Grant No. 2010CD191 to JG); ‘A Hundred Talent Program’ from Chinese Academy of Sciences to PS and ZM, respectively; the three grants to ZM (that is, ‘Top Talents Program in Science and Technology’ from Yunnan Province, ‘Top Talents from Overseas’ from Yunan Province and ‘Innovative Research Initiative of the Synergy between the Natural and Computational Evolutions’ of CAS-Yunan Province). Sequences were deposited in the NCBI Sequence Read Archive (accession number SRA052611).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
ZZ, JG and PS designed the experiments. ZZ, JG, XT and HF generated the data. ZZ, JG, JX, XW, ZM and PS analysed the data. ZZ, ZM and PS wrote the manuscript with inputs from the other members of the team. All authors read and approved the final manuscript.
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Zhang, Z., Geng, J., Tang, X. et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J 8, 881–893 (2014). https://doi.org/10.1038/ismej.2013.185
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.185
Keywords
This article is cited by
-
Correlation analysis of lung mucosa-colonizing bacteria with clinical features reveals metastasis-associated bacterial community structure in non-small cell lung cancer patients
Respiratory Research (2023)
-
Identifying keystone species in microbial communities using deep learning
Nature Ecology & Evolution (2023)
-
Identification of inulin-responsive bacteria in the gut microbiota via multi-modal activity-based sorting
Nature Communications (2023)
-
Administration of probiotics to healthy volunteers: effects on reactivity of intestinal mucosa and systemic leukocytes
BMC Gastroenterology (2022)
-
The impacts of faecal subsampling on microbial compositional profiling
BMC Research Notes (2022)