Abstract
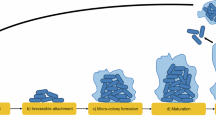
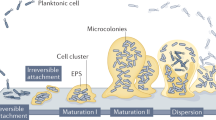
Most studies of biofilm biology have taken a reductionist approach, where single-species biofilms have been extensively investigated. However, biofilms in nature mostly comprise multiple species, where interspecies interactions can shape the development, structure and function of these communities differently from biofilm populations. Hence, a reproducible mixed-species biofilm comprising Pseudomonas aeruginosa, Pseudomonas protegens and Klebsiella pneumoniae was adapted to study how interspecies interactions affect biofilm development, structure and stress responses. Each species was fluorescently tagged to determine its abundance and spatial localization within the biofilm. The mixed-species biofilm exhibited distinct structures that were not observed in comparable single-species biofilms. In addition, development of the mixed-species biofilm was delayed 1–2 days compared with the single-species biofilms. Composition and spatial organization of the mixed-species biofilm also changed along the flow cell channel, where nutrient conditions and growth rate of each species could have a part in community assembly. Intriguingly, the mixed-species biofilm was more resistant to the antimicrobials sodium dodecyl sulfate and tobramycin than the single-species biofilms. Crucially, such community level resilience was found to be a protection offered by the resistant species to the whole community rather than selection for the resistant species. In contrast, community-level resilience was not observed for mixed-species planktonic cultures. These findings suggest that community-level interactions, such as sharing of public goods, are unique to the structured biofilm community, where the members are closely associated with each other.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Anand AA, Vennison SJ, Sankar SG, Prabhu DI, Vasan PT, Raghuraman T et al (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci 10: 107.
Arciola CR, Campoccia D, Gamberini S, Donati ME, Pirini V, Visai L et al (2005). Antibiotic resistance in exopolysaccharide-forming Staphylococcus epidermidis clinical isolates from orthopaedic implant infections. Biomaterials 26: 6530–6535.
Banks MK, Bryers JD . (1991). Bacterial species dominance within a binary culture biofilm. Appl Environ Microbiol 57: 1974–1979.
Breugelmans P, Barken KB, Tolker-Nielsen T, Hofkens J, Dejonghe W, Springael D . (2008). Architecture and spatial organization in a triple-species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiol Ecol 64: 271–282.
Burmolle M, Webb JS, Rao D, Hansen LH, Sorensen SJ, Kjelleberg S . (2006). Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72: 3916–3923.
Chazal PM . (1995). Pollution of modern metalworking fluids containing biocides by pathogenic bacteria in France. Reexamination of chemical treatments accuracy. Eur J Epidemiol 11: 1–7.
Childers BM, Van Laar TA, You T, Clegg S, Leung KP . (2013). MrkD(1P) from Klebsiella pneumoniae IA565 allows for co-existence with Pseudomonas aeruginosa and protection from protease-mediated biofilm detachment. Infect Immun 81: 4112–4120.
Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR et al (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat Meth 2: 443–448.
Choi K-H, Kumar A, Schweizer HP . (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Meth 64: 391–397.
Costerton JW, Stewart PS, Greenberg EP . (1999). Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322.
Cowan SE, Gilbert E, Liepmann D, Keasling JD . (2000). Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl Environ Microbiol 66: 4481–4485.
de Lorenzo V, Timmis KN . (1994). Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235: 386–405.
Fournet-Fayard S, Joly B, Forestier C . (1995). Transformation of wild type Klebsiella pneumoniae with plasmid DNA by electroporation. J Microbiol Meth 24: 49–54.
Hadjantonakis A-K, Macmaster S, Nagy A . (2002). Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol 2: 11.
Hagelueken G, Adams TM, Wiehlmann L, Widow U, Kolmar H, Tummler B et al (2006). The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc Natl Acad Sci USA 103: 7631–7636.
Harrison F . (2007). Microbial ecology of the cystic fibrosis lung. Microbiology 153: 917–923.
Heydorn A, Ersboll BK, Hentzer M, Parsek MR, Givskov M, Molin S . (2000). Experimental reproducibility in flow-chamber biofilms. Microbiology 146 (Pt 10): 2409–2415.
Jackson G, Beyenal H, Rees WM, Lewandowski Z . (2001). Growing reproducible biofilms with respect to structure and viable cell counts. J Microbiol Meth 47: 1–10.
James GA, Beaudette L, Costerton JW . (1995). Interspecies bacterial interactions in biofilms. J Ind Microbiol 15: 257–262.
Jensen PO, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L et al (2007). Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153: 1329–1338.
Korber DR, Lawrence JR, Hendry MJ, Caldwell DE . (1993). Analysis of spatial variability within mot+ and mot− Pseudomonas fluorescens biofilms using representative elements. Biofouling 7: 339–358.
Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT . (2013a). The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ Microbiol 15: 702–715.
Lim YW, Schmieder R, Haynes M, Willner D, Furlan M, Youle M et al (2013b). Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J Cyst Fibros 12: 154–164.
Mah T-FC, O'Toole GA . (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9: 34–39.
Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM . (1999). Aminoglycosides: activity and resistance. Antimicrob Agents Chemother 43: 727–737.
Miller VL, Mekalanos JJ . (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170: 2575–2583.
Moons P, Michiels CW, Aertsen A . (2009). Bacterial interactions in biofilms. Crit Rev Microbiol 35: 157–168.
Nickel JC, Ruseska I, Wright JB, Costerton JW . (1985). Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 27: 619–624.
Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S . (2000). Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ Microbiol 2: 59–68.
Paster BJ, Olsen I, Aas JA, Dewhirst FE . (2006). The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 42: 80–87.
Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer JM, Defago G et al (2011). Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol 34: 180–188.
Rao D, Webb JS, Kjelleberg S . (2005). Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol 71: 1729–1736.
Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD . (2004). Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 42: 5176–5183.
Sauer K, Camper AK . (2001). Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol 183: 6579–6589.
Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG . (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184: 1140–1154.
Schembri MA, Kjaergaard K, Klemm P . (2003). Global gene expression in Escherichia coli biofilms. Mol Microbiol 48: 253–267.
Sternberg C, Tolker-Nielsen T . (2005) Growing and analyzing biofilms in flow cells. John Wiley and Sons, Inc.: Hoboken, New Jersey.
Stoodley P, Wilson S, Hall-Stoodley L, Boyle JD, Lappin-Scott HM, Costerton JW . (2001). Growth and detachment of cell clusters from mature mixed-species biofilms. Appl Environ Microbiol 67: 5608–5613.
Tao W, Evans BG, Yao J, Cooper S, Cornetta K, Ballas CB et al (2007). Enhanced green fluorescent protein is a nearly ideal long-term expression tracer for hematopoietic stem cells, whereas DsRed-express fluorescent protein is not. Stem Cells 25: 670–678.
Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S et al (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864.
Yanisch-Perron C, Vieira J, Messing J . (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33: 103–119.
Yu W, Dodds W, Banks M, Skalsky J, Strauss E . (1995). Optimal staining and sample storage time for direct microscopic enumeration of total and active bacteria in soil with two fluorescent dyes. Appl Environ Microbiol 61: 3367–3372.
Zhang T, Shao M-F, Ye L . (2012). 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6: 1137–1147.
Acknowledgements
Kai Wei Kelvin Lee is supported by the National Research Foundation Singapore under the National Research Foundation (NRF) Environmental and Water Technologies (EWT) PhD Scholarship Programme and administered by the Environment and Water Industry Programme Office (EWI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, K., Periasamy, S., Mukherjee, M. et al. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J 8, 894–907 (2014). https://doi.org/10.1038/ismej.2013.194
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.194
Keywords
This article is cited by
-
Environmental stress mediates groundwater microbial community assembly
Nature Microbiology (2024)
-
Surface-layer protein is a public-good matrix exopolymer for microbial community organisation in environmental anammox biofilms
The ISME Journal (2023)
-
Survival strategies of Listeria monocytogenes to environmental hostile stress: biofilm formation and stress responses
Food Science and Biotechnology (2023)
-
Biotic Interactions in Soil are Underestimated Drivers of Microbial Carbon Use Efficiency
Current Microbiology (2023)
-
Inhibitory effect of ficin on Candida albicans biofilm formation and pre-formed biofilms
BMC Oral Health (2022)