Abstract
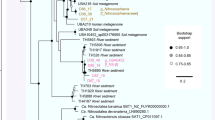
Previous studies based on analysis of amoA, 16S ribosomal RNA or accA gene sequences have established that marine Thaumarchaeota fall into two phylogenetically distinct groups corresponding to shallow- and deep-water clades, but it is not clear how water depth interacts with other environmental factors, including light, temperature and location, to affect this pattern of diversification. Earlier studies focused on single-gene distributions were not able to link phylogenetic structure to other aspects of functional adaptation. Here, we analyzed the genome content of 46 uncultivated single Thaumarchaeota cells sampled from epi- and mesopelagic waters of subtropical, temperate and polar oceans. Phylogenomic analysis showed that populations diverged by depth, as expected, and that mesopelagic populations from different locations were well mixed. Functional analysis showed that some traits, including putative DNA photolyase and catalase genes that may be related to adaptive mechanisms to reduce light-induced damage, were found exclusively in members of the epipelagic clade. Our analysis of partial genomes has thus confirmed the depth differentiation of Thaumarchaeota populations observed previously, consistent with the distribution of putative mechanisms to reduce light-induced damage in shallow- and deep-water populations.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL et al (2012). Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109: 17989–17994.
Biller SJ, Mosier AC, Wells GF, Francis CA . (2012). Global biodiversity of aquatic ammonia-oxidizing Archaea is partitioned by habitat. Front Microbiol 3: 252.
Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR . (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6: e16626.
Church MJ, Wai B, Karl DM, DeLong EF . (2010). Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol 12: 679–688.
Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM . (2003). Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr 48: 1893–1902.
Costa CS, Pezzoni M, Fernández RO, Pizarro RA . (2010). Role of the quorum sensing mechanism in the response of Pseudomonas aeruginosa to lethal and sublethal UVA irradiation. Photochem Photobiol 86: 1334–1342.
Diaz JM, Hansel CM, Voelker BM, Mendes CM, Andeer PF, Zhang T . (2013). Widespread production of extracellular superoxide by heterotrophic bacteria. Science 340: 1223–1226.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing Archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102: 14683–14688.
Goosen N, Moolenaar GF . (2008). Repair of UV damage in bacteria. DNA Repair 7: 353–379.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine. Crenarchaeota. PLoS Biol 4: e95.
Hu A, Jiao N, Zhang R, Yang Z . (2011). Niche partitioning of marine group I Crenarchaeota in the euphotic and upper mesopelagic zones of the East China Sea. Appl Environ Microbiol 77: 7469–7478.
Kalanetra KM, Bano N, Hollibaugh JT . (2009). Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters. Environ Microbiol 11: 2434–2445.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Lanfear R, Calcott B, Ho SYW, Guindon S . (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701.
Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al (2009). The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Luo H, Csűros M, Hughes AL, Moran MA . (2013). Evolution of divergent life history strategies in marine Alphaproteobacteria. mBio 4: e00373–13.
Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI . (2012). Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327: 41–46.
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF . (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9: 1162–1175.
Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA . (2012a). Genome sequence of ‘candidatus Nitrosoarchaeum limnia’ BG20, a low-salinity ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194: 2119–2120.
Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA . (2012b). Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194: 2121–2122.
Nicol GW, Leininger S, Schleper C . (2011). Distribution and activity of ammonia-oxidizing Archaea in natural environments. In Ward BB, Arp DJ, Klotz MJ (eds) Nitrification. ASM Press: Washington, DC, pp 157–178.
Park S-J, Kim J-G, Jung M-Y, Kim S-J, Cha I-T, Kwon K et al (2012a). Draft genome sequence of an ammonia-oxidizing Archaeon, ‘Candidatus Nitrosopumilus koreensis’ AR1, from marine sediment. J Bacteriol 194: 6940–6941.
Park S-J, Kim J-G, Jung M-Y, Kim S-J, Cha I-T, Ghai R et al (2012b). Draft genome sequence of an ammonia-oxidizing Archaeon, ‘Candidatus Nitrosopumilus sediminis’ AR2, from svalbard in the Arctic circle. J Bacteriol 194: 6948–6949.
Parsonage D, Karplus PA, Poole LB . (2008). Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci USA 105: 8209–8214.
Prosser JI, Nicol GW . (2008). Relative contributions of Archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10: 2931–2941.
Rodriguez-Brito B, Rohwer F, Edwards R . (2006). An application of statistics to comparative metagenomics. BMC Bioinformatics 7: 162.
Ronquist F, Huelsenbeck JP . (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Tatusov RL, Koonin EV, Lipman DJ . (1997). A genomic perspective on protein families. Science 278: 631–637.
Wolf Y, Makarova K, Yutin N, Koonin E . (2012). Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7: 46.
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P et al (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103: 12317–12322.
Acknowledgements
We thank the Georgia Advanced Computing Resource Center at the University of Georgia for providing computational resources. This research was funded by grants from NSF (OPP 08-38996; OCE-1232982, EF-826924 and OCE-821374) and the Gordon and Betty Moore Foundation, with sequencing support from the US Department of Energy Joint Genome Institute Community Supported Program grants 2010-77 and 2011-387.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Luo, H., Tolar, B., Swan, B. et al. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J 8, 732–736 (2014). https://doi.org/10.1038/ismej.2013.202
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.202
Keywords
This article is cited by
-
Diversity and salinity adaptations of ammonia oxidizing archaea in three estuaries of China
Applied Microbiology and Biotechnology (2023)
-
Phylotype resolved spatial variation and association patterns of planktonic Thaumarchaeota in eastern Chinese marginal seas
Marine Life Science & Technology (2023)
-
Phylogenetic divergence and adaptation of Nitrososphaeria across lake depths and freshwater ecosystems
The ISME Journal (2022)
-
Marine ammonia-oxidising archaea and bacteria occupy distinct iron and copper niches
ISME Communications (2021)
-
Novel insights into the Thaumarchaeota in the deepest oceans: their metabolism and potential adaptation mechanisms
Microbiome (2020)